Metadherin Is a Prognostic Apoptosis Modulator in Mesothelioma Induced via NF-κB-Mediated Signaling
- PMID: 31054476
- PMCID: PMC6500914
- DOI: 10.1016/j.tranon.2019.03.005
Metadherin Is a Prognostic Apoptosis Modulator in Mesothelioma Induced via NF-κB-Mediated Signaling
Abstract
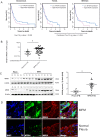
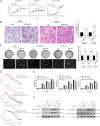
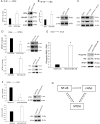
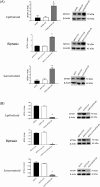
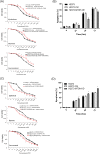
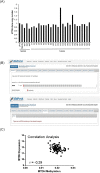
Therapies against malignant pleural mesothelioma (MPM) have yielded disappointing results, in part, because pathologic mechanisms remain obscure. In searching for rational molecular targets, we identified metadherin (MTDH), a multifunctional gene associated with several tumor types but previously unrecognized in MPM. Cox proportional hazards regression analysis delineated associations between higher MTDH expression and lower patient survival from three independent MPM cohorts (n = 349 patients). Through in vitro assays with overexpression and downregulation constructs in MPM cells, we characterized the role of MTDH. We confirmed in vivo the phenotype of altered MTDH expression in a murine xenograft model. Transcriptional regulators of MTDH were identified by chromatin immunoprecipitation. Overexpression of both MTDH mRNA (12-fold increased) and protein levels was observed in tumor tissues. MTDH stable overexpression significantly augmented proliferation, invasiveness, colony formation, chemoresistance, and an antiapoptosis phenotype, while its suppression showed opposite effects in MPM cells. Interestingly, NF-κB and c-Myc (in a feed-forward loop motif) contributed to modulating MTDH expression. Knockdown of MTDH expression profoundly retarded xenograft tumor growth. Thus, our findings support the notion that MTDH integrates upstream signals from certain transcription factors and mediates pathogenic interactions contributing to MPM traits. MTDH represents a new MPM-associated gene that can contribute to insights of MPM biology and, as such, suggest other treatment strategies.
Published by Elsevier Inc.
Figures







References
-
- Beckett P, Edwards J, Fennell D, Hubbard R, Woolhouse I, Peake MD. Demographics, management and survival of patients with malignant pleural mesothelioma in the National Lung Cancer Audit in England and Wales. Lung Cancer. 2015;88:344–348. - PubMed
-
- Zalcman G, Mazieres J, Margery J, Greillier L, Audigier-Valette C, Moro-Sibilot D, Molinier O, Corre R, Monnet I, Gounant V. Bevacizumab for newly diagnosed pleural mesothelioma in the Mesothelioma Avastin Cisplatin Pemetrexed Study (MAPS): a randomised, controlled, open-label, phase 3 trial. Lancet. 2016;387:1405–1414. - PubMed
-
- Bueno R, Stawiski EW, Goldstein LD, Durinck S, De Rienzo A, Modrusan Z, Gnad F, Nguyen TT, Jaiswal BS, Chirieac LR. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48:407–416. - PubMed
-
- Li J, Zhang N, Song LB, Liao WT, Jiang LL, Gong LY, Wu J, Yuan J, Zhang HZ, Zeng MS. Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res. 2008;14:3319–3326. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

