A brief history of bird flu
- PMID: 31056053
- PMCID: PMC6553608
- DOI: 10.1098/rstb.2018.0257
A brief history of bird flu
Abstract
In 1918, a strain of influenza A virus caused a human pandemic resulting in the deaths of 50 million people. A century later, with the advent of sequencing technology and corresponding phylogenetic methods, we know much more about the origins, evolution and epidemiology of influenza epidemics. Here we review the history of avian influenza viruses through the lens of their genetic makeup: from their relationship to human pandemic viruses, starting with the 1918 H1N1 strain, through to the highly pathogenic epidemics in birds and zoonoses up to 2018. We describe the genesis of novel influenza A virus strains by reassortment and evolution in wild and domestic bird populations, as well as the role of wild bird migration in their long-range spread. The emergence of highly pathogenic avian influenza viruses, and the zoonotic incursions of avian H5 and H7 viruses into humans over the last couple of decades are also described. The threat of a new avian influenza virus causing a human pandemic is still present today, although control in domestic avian populations can minimize the risk to human health. This article is part of the theme issue 'Modelling infectious disease outbreaks in humans, animals and plants: approaches and important themes'. This issue is linked with the subsequent theme issue 'Modelling infectious disease outbreaks in humans, animals and plants: epidemic forecasting and control'.
Keywords: avian influenza virus; epidemiology; pandemic; phylogenetics; zoonotic.
Conflict of interest statement
We have no competing interests
Figures
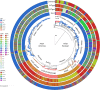
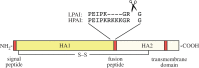
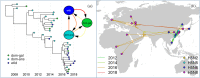
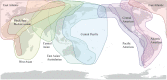
References
-
- King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. 2012. Virus taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses, pp. 749–761. Amsterdam, The Netherlands: Elsevier; ( 10.1016/B978-0-12-384684-6.00136-1) - DOI
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
