Analysis of Subtelomeric REXTAL Assemblies Using QUAST
- PMID: 31056507
- PMCID: PMC6940546
- DOI: 10.1109/TCBB.2019.2913845
Analysis of Subtelomeric REXTAL Assemblies Using QUAST
Abstract
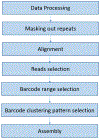
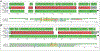
Genomic regions of high segmental duplication content and/or structural variation have led to gaps and misassemblies in the human reference sequence, and are refractory to assembly from whole-genome short-read datasets. Human subtelomere regions are highly enriched in both segmental duplication content and structural variations, and as a consequence are both impossible to assemble accurately and highly variable from individual to individual. Recently, we developed a pipeline for improved region-specific assembly called Regional Extension of Assemblies Using Linked-Reads (REXTAL). In this study, we evaluate REXTAL and genome-wide assembly (Supernova) approaches on 10X Genomics linked-reads data sets partitioned and barcoded using the Gel Bead in Emulsion (GEM) microfluidic method. Our results describe the accuracy and relative performance of these two approaches using the reference-based assessment module of QUAST. We show that REXTAL dramatically outperforms the Supernova whole genome assembler in subtelomeric segmental duplication regions, and results in highly accurate assemblies. Nearly all of the REXTAL "misassemblies" identified using default QUAST parameters simply pinpoint locations of tandem repeat arrays in the reference sequence where the repeat array length differs from that in the cognate REXTAL assembly by 1000 bp.
Figures

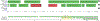

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

