Finding a Compatible Partner: Self-Incompatibility in European Pear (Pyrus communis); Molecular Control, Genetic Determination, and Impact on Fertilization and Fruit Set
- PMID: 31057563
- PMCID: PMC6477101
- DOI: 10.3389/fpls.2019.00407
Finding a Compatible Partner: Self-Incompatibility in European Pear (Pyrus communis); Molecular Control, Genetic Determination, and Impact on Fertilization and Fruit Set
Abstract
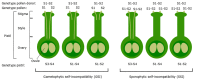
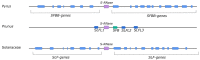
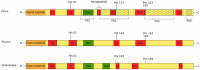
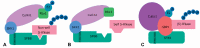
Pyrus species display a gametophytic self-incompatibility (GSI) system that actively prevents fertilization by self-pollen. The GSI mechanism in Pyrus is genetically controlled by a single locus, i.e., the S-locus, which includes at least two polymorphic and strongly linked S-determinant genes: a pistil-expressed S-RNase gene and a number of pollen-expressed SFBB genes (S-locus F-Box Brothers). Both the molecular basis of the SI mechanism and its functional expression have been widely studied in many Rosaceae fruit tree species with a particular focus on the characterization of the elusive SFBB genes and S-RNase alleles of economically important cultivars. Here, we discuss recent advances in the understanding of GSI in Pyrus and provide new insights into the mechanisms of GSI breakdown leading to self-fertilization and fruit set. Molecular analysis of S-genes in several self-compatible Pyrus cultivars has revealed mutations in both pistil- or pollen-specific parts that cause breakdown of self-incompatibility. This has significantly contributed to our understanding of the molecular and genetic mechanisms that underpin self-incompatibility. Moreover, the existence and development of self-compatible mutants open new perspectives for pear production and breeding. In this framework, possible consequences of self-fertilization on fruit set, development, and quality in pear are also reviewed.
Keywords: Pyrus communis; S-RNase; SFBB; fertilization; fruit set; gametophytic self-incompatibility.
Figures

References
-
- Adachi Y., Komori S., Hoshikawa Y., Tanaka N., Abe K., Bessho H., et al. (2009). Characteristics of fruiting and pollen tube growth of apple autotetraploid cultivars showing self-compatibility. J. Japan. Soc. Hort. Sci. 78, 402–409. 10.2503/jjshs1.78.402 - DOI
-
- Anderson M. A., Cornish E. C., Mau S.-L., Williams E. G., Hoggart R., Atkinson A., et al. (1986). Cloning of CDNA for a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata. Nature 321, 38–44. 10.1038/321038a0 - DOI
-
- Atwell B. J., Kriedemann P. E., Turnbull C. G. N. (1999). Plants in action: Adaptation in nature, performance in cultivation. (Australia: Macmillan Education; ).
Publication types
LinkOut - more resources
Full Text Sources

