Classification and Interactions of LRR Receptors and Co-receptors Within the Arabidopsis Plasma Membrane - An Overview
- PMID: 31057579
- PMCID: PMC6477698
- DOI: 10.3389/fpls.2019.00472
Classification and Interactions of LRR Receptors and Co-receptors Within the Arabidopsis Plasma Membrane - An Overview
Abstract
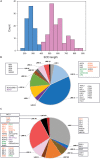
Receptor kinases (RK) constitute the largest protein kinase family in plants. In particular, members of the leucine-rich repeat-receptor kinases (LRR-RKs) are involved in the perception of various signals at the plasma membrane. Experimental evidence over the past years revealed a conserved activation mechanism through ligand-inducible heterodimer formation: a ligand is recognized by a receptor kinase with a large extracellular domain (ECD). This ligand binding receptor directly interacts with a so-called co-receptor with a small ECD for ligand fixation and kinase activation. A large proportion of LRR-RKs is functionally still uncharacterized and the dynamic complexity of the plasma membrane makes it difficult to precisely define receptor kinase heterodimer pairs and their functions. In this review, we give an overview of the current knowledge of LRR receptor and co-receptor functions. We use ECD lengths to classify the LRR receptor kinase family and describe different interaction properties of ligand-binding receptors and their respective co-receptor from a network perspective.
Keywords: LRR-RKs; co-receptor; extracellular domain lengths; ligand-perceiving; plasma membrane.
Figures

References
LinkOut - more resources
Full Text Sources
Miscellaneous

