MicroRNA‑93 regulates angiogenesis in peripheral arterial disease by targeting CDKN1A
- PMID: 31059098
- PMCID: PMC6522868
- DOI: 10.3892/mmr.2019.10196
MicroRNA‑93 regulates angiogenesis in peripheral arterial disease by targeting CDKN1A
Abstract
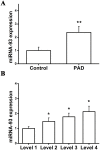
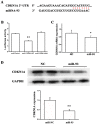
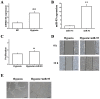
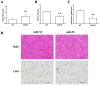
MicroRNAs (miRNAs) are considered to be critical mediators of gene expression with respect to tumor progression, although their role in ischemia‑induced angiogenesis is poorly characterized, including in peripheral arterial disease (PAD). Furthermore, the underlying mechanism of action of specific miRNAs in PAD remains unknown. Reverse transcription‑quantitative polymerase chain reaction analysis revealed that microRNA‑93 (miR‑93) was significantly upregulated in patients with PAD and in the EA.hy926 endothelial cells in response to hypoxia. Additionally, miRNA (miR)‑93 promoted angiogenesis by enhancing proliferation, migration and tube formation. Cyclin dependent kinase inhibitor 1A (CDKN1A), verified as a potential target gene of miR‑93, was inhibited by overexpressed miR‑93 at the protein and mRNA expression levels. Furthermore, a hind‑limb ischemia model served to evaluate the role of miR‑93 in angiogenesis in vivo, and the results demonstrated that miR‑93 overexpression enhanced capillary density and perfusion recovery from hind‑limb ischemia. Taken together, miR‑93 was indicated to be a promising target for pharmacological regulation to promote angiogenesis, and the miR‑93/CDKN1A pathway may function as a novel therapeutic approach in PAD.
Figures
Similar articles
-
miR-548j-5p regulates angiogenesis in peripheral artery disease.Sci Rep. 2022 Jan 17;12(1):838. doi: 10.1038/s41598-022-04770-6. Sci Rep. 2022. PMID: 35039547 Free PMC article.
-
A MicroRNA93-Interferon Regulatory Factor-9-Immunoresponsive Gene-1-Itaconic Acid Pathway Modulates M2-Like Macrophage Polarization to Revascularize Ischemic Muscle.Circulation. 2017 Jun 13;135(24):2403-2425. doi: 10.1161/CIRCULATIONAHA.116.025490. Epub 2017 Mar 29. Circulation. 2017. PMID: 28356443 Free PMC article.
-
Human Umbilical Cord-Derived Mesenchymal Stem Cells Relieve Hind Limb Ischemia by Promoting Angiogenesis in Mice.Stem Cells Dev. 2019 Oct 15;28(20):1384-1397. doi: 10.1089/scd.2019.0115. Epub 2019 Sep 18. Stem Cells Dev. 2019. PMID: 31407635
-
MiRNAs in peripheral artery disease - something gripping this way comes.Vasa. 2014 May;43(3):163-70. doi: 10.1024/0301-1526/a000345. Vasa. 2014. PMID: 24797047 Review.
-
Role of microRNAs in peripheral artery disease (review).Mol Med Rep. 2012 Oct;6(4):695-700. doi: 10.3892/mmr.2012.978. Epub 2012 Jul 5. Mol Med Rep. 2012. PMID: 22767222 Review.
Cited by
-
miR-548j-5p regulates angiogenesis in peripheral artery disease.Sci Rep. 2022 Jan 17;12(1):838. doi: 10.1038/s41598-022-04770-6. Sci Rep. 2022. PMID: 35039547 Free PMC article.
-
Single-cell sequencing reveals the existence of fetal vascular endothelial stem cell-like cells in mouse liver.Stem Cell Res Ther. 2023 Aug 30;14(1):227. doi: 10.1186/s13287-023-03460-y. Stem Cell Res Ther. 2023. PMID: 37649114 Free PMC article.
-
Effect of X-rays on transcript expression of rat brain microvascular endothelial cells: role of calcium signaling in X-ray-induced endothelium damage.Biosci Rep. 2020 Apr 30;40(4):BSR20193760. doi: 10.1042/BSR20193760. Biosci Rep. 2020. PMID: 32285918 Free PMC article.
-
Relationships between Indicators of Lower Extremity Artery Disease and miRNA Expression in Peripheral Blood Mononuclear Cells.J Clin Med. 2022 Mar 15;11(6):1619. doi: 10.3390/jcm11061619. J Clin Med. 2022. PMID: 35329950 Free PMC article.
-
Dysregulated Genes, MicroRNAs, Biological Pathways, and Gastrocnemius Muscle Fiber Types Associated With Progression of Peripheral Artery Disease: A Preliminary Analysis.J Am Heart Assoc. 2022 Nov;11(21):e023085. doi: 10.1161/JAHA.121.023085. Epub 2022 Oct 27. J Am Heart Assoc. 2022. PMID: 36300658 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

