Ipomoeassin F Binds Sec61α to Inhibit Protein Translocation
- PMID: 31059257
- PMCID: PMC6627486
- DOI: 10.1021/jacs.8b13506
Ipomoeassin F Binds Sec61α to Inhibit Protein Translocation
Abstract
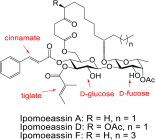
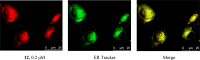
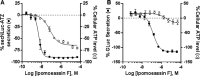
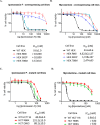
Ipomoeassin F is a potent natural cytotoxin that inhibits growth of many tumor cell lines with single-digit nanomolar potency. However, its biological and pharmacological properties have remained largely unexplored. Building upon our earlier achievements in total synthesis and medicinal chemistry, we used chemical proteomics to identify Sec61α (protein transport protein Sec61 subunit alpha isoform 1), the pore-forming subunit of the Sec61 protein translocon, as a direct binding partner of ipomoeassin F in living cells. The interaction is specific and strong enough to survive lysis conditions, enabling a biotin analogue of ipomoeassin F to pull down Sec61α from live cells, yet it is also reversible, as judged by several experiments including fluorescent streptavidin staining, delayed competition in affinity pulldown, and inhibition of TNF biogenesis after washout. Sec61α forms the central subunit of the ER protein translocation complex, and the binding of ipomoeassin F results in a substantial, yet selective, inhibition of protein translocation in vitro and a broad ranging inhibition of protein secretion in live cells. Lastly, the unique resistance profile demonstrated by specific amino acid single-point mutations in Sec61α provides compelling evidence that Sec61α is the primary molecular target of ipomoeassin F and strongly suggests that the binding of this natural product to Sec61α is distinctive. Therefore, ipomoeassin F represents the first plant-derived, carbohydrate-based member of a novel structural class that offers new opportunities to explore Sec61α function and to further investigate its potential as a therapeutic target for drug discovery.
Conflict of interest statement
The authors declare no competing financial interest.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

