Arboviral screening of invasive Aedes species in northeastern Turkey: West Nile virus circulation and detection of insect-only viruses
- PMID: 31059502
- PMCID: PMC6522068
- DOI: 10.1371/journal.pntd.0007334
Arboviral screening of invasive Aedes species in northeastern Turkey: West Nile virus circulation and detection of insect-only viruses
Abstract
Background: The recent reports of Aedes aegypti and Ae. albopictus populations in Turkey, in parallel with the territorial expansion identified in several surrounding countries, have raised concerns about the establishment and re-establishment of these invasive Aedes mosquitoes in Turkey. This cross-sectional study was performed to detect Aedes aegypti and Ae. albopictus in regions of recent incursions, and screen for viral pathogens known to be transmitted elsewhere by these species.
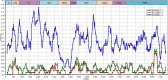
Methodology: Mosquitoes were collected at several locations in Artvin, Rize and Trabzon provinces of the Black Sea region during 2016-2017, identified morphologically, pooled and analyzed via generic or specific nucleic acid amplification assays. Viruses in positive pools were identified by product sequencing, cell culture inoculation and next generation sequencing (NGS) in selected specimens.
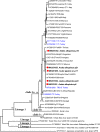
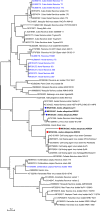
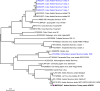
Principal findings: The study group comprised 791 specimens. Aedes albopictus was the most abundant species in all locations (89.6%), followed by Ae. aegypti (7.8%) and Culex pipiens (2.5%). Mosquitoes were screened for viruses in 65 pools where fifteen (23.1%) were reactive. The infecting strains was identified as West Nile virus (WNV) in 5 pools (7.7%) with Ae. albopictus or Cx. pipiens mosquitoes. The obtained WNV sequences phylogenetically grouped with local and global lineage 1 clade 1a viruses. In 4 (6.2%) and 6 (9.2%) pools, respectively, cell fusing agent virus (CFAV) and Aedes flavivirus (AEFV) sequences were characterized. NGS provided a near-complete AEFV genome in a pool of Ae. albopictus. The strain is provisionally called "AEFV-Turkey", and functional analysis of the genome revealed several conserved motifs and regions associated with virus replication. Merida-like virus Turkey (MERDLVT), a recently-described novel rhabdovirus, was also co-detected in a Cx. pipiens pool also positive for WNV.
Conclusions/significance: Invasive Aedes mosquitoes are established in certain locations of northeastern Turkey. Herein we conclusively show the role of these species in WNV circulation in the region. Biosurveillance is imperative to monitor the spread of these species further into Asia Minor and to detect possible introduction of pathogens.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

