Targeted amplification of a sequence of interest in artificial chromosome in mammalian cells
- PMID: 31062017
- PMCID: PMC6582328
- DOI: 10.1093/nar/gkz343
Targeted amplification of a sequence of interest in artificial chromosome in mammalian cells
Abstract
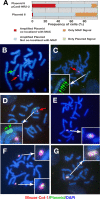
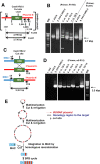
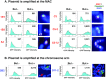
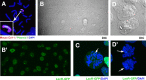
A plasmid with a replication initiation region (IR) and a matrix attachment region (MAR) initiates gene amplification in mammalian cells at a random chromosomal location. A mouse artificial chromosome (MAC) vector can stably carry a large genomic region. In this study we combined these two technologies with the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease (Cas)9 strategy to achieve targeted amplification of a sequence of interest. We previously showed that the IR/MAR plasmid was amplified up to the extrachromosomal tandem repeat; here we demonstrate that cleavage of these tandem plasmids and MAC by Cas9 facilitates homologous recombination between them. The plasmid array on the MAC could be further extended to form a ladder structure with high gene expression by a breakage-fusion-bridge cycle involving breakage at mouse major satellites. Amplification of genes on the MAC has the advantage that the MAC can be transferred between cells. We visualized the MAC in live cells by amplifying the lactose operator array on the MAC in cells expressing lactose repressor-green fluorescent protein fusion protein. This targeted amplification strategy is in theory be applicable to any sequence at any chromosomal site, and provides a novel tool for animal cell technology.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
How a replication origin and matrix attachment region accelerate gene amplification under replication stress in mammalian cells.PLoS One. 2014 Jul 25;9(7):e103439. doi: 10.1371/journal.pone.0103439. eCollection 2014. PLoS One. 2014. PMID: 25061979 Free PMC article.
-
Amplification of a transgene within a long array of replication origins favors higher gene expression in animal cells.PLoS One. 2017 Apr 12;12(4):e0175585. doi: 10.1371/journal.pone.0175585. eCollection 2017. PLoS One. 2017. PMID: 28403180 Free PMC article.
-
Dissection of the beta-globin replication-initiation region reveals specific requirements for replicator elements during gene amplification.PLoS One. 2013 Oct 4;8(10):e77350. doi: 10.1371/journal.pone.0077350. eCollection 2013. PLoS One. 2013. PMID: 24124615 Free PMC article.
-
Extrachromosomal double minutes and chromosomal homogeneously staining regions as probes for chromosome research.Cytogenet Genome Res. 2009;124(3-4):312-26. doi: 10.1159/000218135. Epub 2009 Jun 25. Cytogenet Genome Res. 2009. PMID: 19556783 Review.
-
Advances in therapeutic CRISPR/Cas9 genome editing.Transl Res. 2016 Feb;168:15-21. doi: 10.1016/j.trsl.2015.09.008. Epub 2015 Sep 26. Transl Res. 2016. PMID: 26470680 Review.
Cited by
-
An efficient protein production system via gene amplification on a human artificial chromosome and the chromosome transfer to CHO cells.Sci Rep. 2019 Nov 18;9(1):16954. doi: 10.1038/s41598-019-53116-2. Sci Rep. 2019. PMID: 31740706 Free PMC article.
-
SIRT1 stabilizes extrachromosomal gene amplification and contributes to repeat-induced gene silencing.J Biol Chem. 2021 Jan-Jun;296:100356. doi: 10.1016/j.jbc.2021.100356. Epub 2021 Feb 2. J Biol Chem. 2021. PMID: 33539925 Free PMC article.
-
Revisiting characteristics of oncogenic extrachromosomal DNA as mobile enhancers on neuroblastoma and glioma cancers.Cancer Cell Int. 2022 May 25;22(1):200. doi: 10.1186/s12935-022-02617-8. Cancer Cell Int. 2022. PMID: 35614494 Free PMC article. Review.
-
Gene Amplification and the Extrachromosomal Circular DNA.Genes (Basel). 2021 Sep 28;12(10):1533. doi: 10.3390/genes12101533. Genes (Basel). 2021. PMID: 34680928 Free PMC article. Review.
References
-
- Wahl G.M. The importance of circular DNA in mammalian gene amplification. Cancer Res. 1989; 49:1333–1340. - PubMed
-
- Albertson D.G. Gene amplification in cancer. Trends Genet. 2006; 22:447–455. - PubMed
-
- Shimizu N. Extrachromosomal double minutes and chromosomal homogeneously staining regions as probes for chromosome research. Cytogenet. Genome Res. 2009; 124:312–326. - PubMed
-
- Shimizu N., Miura Y., Sakamoto Y., Tsutsui K.. Plasmids with a mammalian replication origin and a matrix attachment region initiate the event similar to gene amplification. Cancer Res. 2001; 61:6987–6990. - PubMed
-
- Shimizu N., Hashizume T., Shingaki K., Kawamoto J.K.. Amplification of plasmids containing a mammalian replication initiation region is mediated by controllable conflict between replication and transcription. Cancer Res. 2003; 63:5281–5290. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

