RhoA regulates translation of the Nogo-A decoy SPARC in white matter-invading glioblastomas
- PMID: 31062076
- PMCID: PMC6660512
- DOI: 10.1007/s00401-019-02021-z
RhoA regulates translation of the Nogo-A decoy SPARC in white matter-invading glioblastomas
Abstract
Glioblastomas strongly invade the brain by infiltrating into the white matter along myelinated nerve fiber tracts even though the myelin protein Nogo-A prevents cell migration by activating inhibitory RhoA signaling. The mechanisms behind this long-known phenomenon remained elusive so far, precluding a targeted therapeutic intervention. This study demonstrates that the prevalent activation of AKT in gliomas increases the ER protein-folding capacity and enables tumor cells to utilize a side effect of RhoA activation: the perturbation of the IRE1α-mediated decay of SPARC mRNA. Once translation is initiated, glioblastoma cells rapidly secrete SPARC to block Nogo-A from inhibiting migration via RhoA. By advanced ultramicroscopy for studying single-cell invasion in whole, undissected mouse brains, we show that gliomas require SPARC for invading into white matter structures. SPARC depletion reduces tumor dissemination that significantly prolongs survival and improves response to cytostatic therapy. Our finding of a novel RhoA-IRE1 axis provides a druggable target for interfering with SPARC production and underscores its therapeutic value.
Keywords: AKT; ENTPD5; Glioblastoma; IRE1α; Invasion; Nogo-A; Post-transcriptional regulation; RhoA; SPARC; White matter.
Conflict of interest statement
GR has received research Grants from Roche and Merck Serono.
Figures
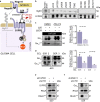
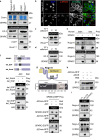

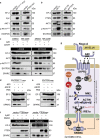
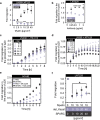
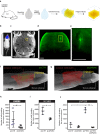
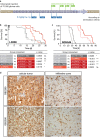
References
Publication types
MeSH terms
Substances
Grants and funding
- Junior Group Leader Award/CHS Stiftung/International
- 01ZX1401A/Bundesministerium für Bildung und Forschung/International
- 01ZX1401C/Bundesministerium für Bildung und Forschung/International
- 01ZX1401B/Bundesministerium für Bildung und Forschung/International
- 01ZX1401/Bundesministerium für Bildung und Forschung/International
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

