Computational 3D histological phenotyping of whole zebrafish by X-ray histotomography
- PMID: 31063133
- PMCID: PMC6559789
- DOI: 10.7554/eLife.44898
Computational 3D histological phenotyping of whole zebrafish by X-ray histotomography
Abstract
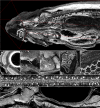
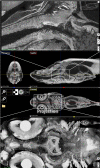
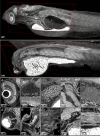
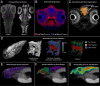
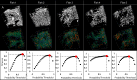
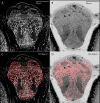
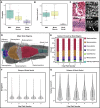
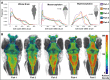
Organismal phenotypes frequently involve multiple organ systems. Histology is a powerful way to detect cellular and tissue phenotypes, but is largely descriptive and subjective. To determine how synchrotron-based X-ray micro-tomography (micro-CT) can yield 3-dimensional whole-organism images suitable for quantitative histological phenotyping, we scanned whole zebrafish, a small vertebrate model with diverse tissues, at ~1 micron voxel resolutions. Micro-CT optimized for cellular characterization (histotomography) allows brain nuclei to be computationally segmented and assigned to brain regions, and cell shapes and volumes to be computed for motor neurons and red blood cells. Striking individual phenotypic variation was apparent from color maps of computed densities of brain nuclei. Unlike histology, the histotomography also allows the study of 3-dimensional structures of millimeter scale that cross multiple tissue planes. We expect the computational and visual insights into 3D cell and tissue architecture provided by histotomography to be useful for reference atlases, hypothesis generation, comprehensive organismal screens, and diagnostics.
Keywords: 3D histology; 3D imaging; cell density; cell shape; computational phenomics; developmental biology; micro-CT; phenotypic variation; tissue architecture; volumetric histology; whole-organism phenomics; zebrafish.
© 2019, Ding et al.
Conflict of interest statement
YD, DV, MY, SK, AL, DC, PV, XX, JC, VC, KA, YW, XX, FD, Dv, PL, KC No competing interests declared
Figures


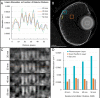
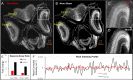

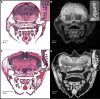
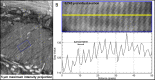

Comment in
- doi: 10.7554/eLife.48340
References
-
- Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, Grieder FB, Heintz N, Hicks G, Insel TR, Joyner A, Koller BH, Lloyd KC, Magnuson T, Moore MW, Nagy A, Pollock JD, Roses AD, Sands AT, Seed B, Skarnes WC, Snoddy J, Soriano P, Stewart DJ, Stewart F, Stillman B, Varmus H, Varticovski L, Verma IM, Vogt TF, von Melchner H, Witkowski J, Woychik RP, Wurst W, Yancopoulos GD, Young SG, Zambrowicz B. The knockout mouse project. Nature genetics. 2004;36:921–924. doi: 10.1038/ng0904-921. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
- Pilot award funding/Huck Institutes of the Life Sciences/International
- Pilot award funding/Jake Gittlen Memorial Golf Tournament/International
- R01 CA131956/CA/NCI NIH HHS/United States
- R24-OD018559/NIH Office of the Director/International
- Penn State Zebrafish Functional Genomics Core/Pennsylvania Tobacco Fund/International
- Pilot award funding/Institute for Cyber Science, PSU/International
- G20 OD016619/OD/NIH HHS/United States
- R24 RR017441/RR/NCRR NIH HHS/United States
- R24 OD018559/OD/NIH HHS/United States
- R01 HD040179/HD/NICHD NIH HHS/United States
- VIrtual Slide Scanner/Huck Institutes of the Life Sciences/International
- R24-RR017441/NH/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

