Biodegradation of Tetralin: Genomics, Gene Function and Regulation
- PMID: 31064110
- PMCID: PMC6563040
- DOI: 10.3390/genes10050339
Biodegradation of Tetralin: Genomics, Gene Function and Regulation
Abstract
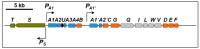
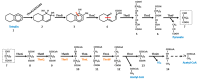
Tetralin (1,2,3,4-tetrahydonaphthalene) is a recalcitrant compound that consists of an aromatic and an alicyclic ring. It is found in crude oils, produced industrially from naphthalene or anthracene, and widely used as an organic solvent. Its toxicity is due to the alteration of biological membranes by its hydrophobic character and to the formation of toxic hydroperoxides. Two unrelated bacteria, Sphingopyxis granuli strain TFA and Rhodococcus sp. strain TFB were isolated from the same niche as able to grow on tetralin as the sole source of carbon and energy. In this review, we provide an overview of current knowledge on tetralin catabolism at biochemical, genetic and regulatory levels in both strains. Although they share the same biodegradation strategy and enzymatic activities, no evidences of horizontal gene transfer between both bacteria have been found. Moreover, the regulatory elements that control the expression of the gene clusters are completely different in each strain. A special consideration is given to the complex regulation discovered in TFA since three regulatory systems, one of them involving an unprecedented communication between the catabolic pathway and the regulatory elements, act together at transcriptional and posttranscriptional levels to optimize tetralin biodegradation gene expression to the environmental conditions.
Keywords: Rhodococcus sp. strain TFB; Sphingopyxis granuli strain TFA; carbon catabolite repression; redox proteins; tetralin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Molecular and biochemical characterization of the tetralin degradation pathway in Rhodococcus sp. strain TFB.Microb Biotechnol. 2009 Mar;2(2):262-73. doi: 10.1111/j.1751-7915.2009.00086.x. Microb Biotechnol. 2009. PMID: 21261920 Free PMC article.
-
SuhB, a small non-coding RNA involved in catabolite repression of tetralin degradation genes in Sphingopyxis granuli strain TFA.Environ Microbiol. 2018 Oct;20(10):3671-3683. doi: 10.1111/1462-2920.14360. Epub 2018 Sep 14. Environ Microbiol. 2018. PMID: 30033661
-
Redox proteins of hydroxylating bacterial dioxygenases establish a regulatory cascade that prevents gratuitous induction of tetralin biodegradation genes.Sci Rep. 2016 Mar 31;6:23848. doi: 10.1038/srep23848. Sci Rep. 2016. PMID: 27030382 Free PMC article.
-
Genome analysis and -omics approaches provide new insights into the biodegradation potential of Rhodococcus.Appl Microbiol Biotechnol. 2019 Feb;103(3):1069-1080. doi: 10.1007/s00253-018-9539-7. Epub 2018 Dec 15. Appl Microbiol Biotechnol. 2019. PMID: 30554387 Review.
-
Biodegradation and Rhodococcus--masters of catabolic versatility.Curr Opin Biotechnol. 2005 Jun;16(3):282-90. doi: 10.1016/j.copbio.2005.04.007. Curr Opin Biotechnol. 2005. PMID: 15961029 Review.
Cited by
-
An improved genome editing system for Sphingomonadaceae.Access Microbiol. 2024 May 13;6(5):000755.v3. doi: 10.1099/acmi.0.000755.v3. eCollection 2024. Access Microbiol. 2024. PMID: 38868378 Free PMC article.
-
Special Issue: Genetics of Biodegradation and Bioremediation.Genes (Basel). 2020 Apr 17;11(4):441. doi: 10.3390/genes11040441. Genes (Basel). 2020. PMID: 32316688 Free PMC article.
-
Understanding the metabolism of the tetralin degrader Sphingopyxis granuli strain TFA through genome-scale metabolic modelling.Sci Rep. 2020 May 26;10(1):8651. doi: 10.1038/s41598-020-65258-9. Sci Rep. 2020. PMID: 32457330 Free PMC article.
-
The functional differences between paralogous regulators define the control of the general stress response in Sphingopyxis granuli TFA.Environ Microbiol. 2022 Apr;24(4):1918-1931. doi: 10.1111/1462-2920.15907. Epub 2022 Jan 27. Environ Microbiol. 2022. PMID: 35049124 Free PMC article.
-
Identification of two fnr genes and characterisation of their role in the anaerobic switch in Sphingopyxis granuli strain TFA.Sci Rep. 2020 Dec 3;10(1):21019. doi: 10.1038/s41598-020-77927-w. Sci Rep. 2020. PMID: 33273546 Free PMC article.
References
-
- Sikkema J., de Bont J.A. Isolation and initial characterization of bacteria growing on tetralin. Biodegradation. 1991;2:15–23. doi: 10.1007/BF00122421. - DOI

