TFIIE orchestrates the recruitment of the TFIIH kinase module at promoter before release during transcription
- PMID: 31064989
- PMCID: PMC6504876
- DOI: 10.1038/s41467-019-10131-1
TFIIE orchestrates the recruitment of the TFIIH kinase module at promoter before release during transcription
Abstract
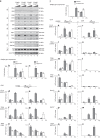
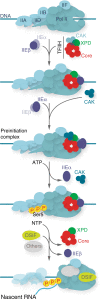
In eukaryotes, the general transcription factors TFIIE and TFIIH assemble at the transcription start site with RNA Polymerase II. However, the mechanism by which these transcription factors incorporate the preinitiation complex and coordinate their action during RNA polymerase II transcription remains elusive. Here we show that the TFIIEα and TFIIEβ subunits anchor the TFIIH kinase module (CAK) within the preinitiation complex. In addition, we show that while RNA polymerase II phosphorylation and DNA opening occur, CAK and TFIIEα are released from the promoter. This dissociation is impeded by either ATP-γS or CDK7 inhibitor THZ1, but still occurs when XPB activity is abrogated. Finally, we show that the Core-TFIIH and TFIIEβ are subsequently removed, while elongation factors such as DSIF are recruited. Remarkably, these early transcriptional events are affected by TFIIE and TFIIH mutations associated with the developmental disorder, trichothiodystrophy.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Maldonado E, Ha I, Cortes P, Weis L, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II: role of transcription factors IIA, IID, and IIB during formation of a transcription-competent complex. Mol. Cell. Biol. 1990;10:6335–6347. doi: 10.1128/MCB.10.12.6335. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- INCA-PLBIO17-043/Institut National Du Cancer (French National Cancer Institute)/International
- 130607082/Fondation ARC pour la Recherche sur le Cancer (ARC Foundation for Cancer Research)/International
- (Equipe labélisée 2019)/Ligue Contre le Cancer/International
- PICS-6824/Centre National de la Recherche Scientifique (National Center for Scientific Research)/International
LinkOut - more resources
Full Text Sources
Research Materials

