Induced Pluripotent Stem Cell Derivation and Ex Vivo Gene Correction Using a Mucopolysaccharidosis Type 1 Disease Mouse Model
- PMID: 31065277
- PMCID: PMC6466856
- DOI: 10.1155/2019/6978303
Induced Pluripotent Stem Cell Derivation and Ex Vivo Gene Correction Using a Mucopolysaccharidosis Type 1 Disease Mouse Model
Abstract
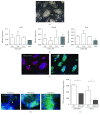
Mucopolysaccharidosis type 1 (MPS-1), also known as Hurler's disease, is a congenital metabolic disorder caused by a mutation in the alpha-L-iduronidase (IDUA) gene, which results in the loss of lysosomal enzyme function for the degradation of glycosaminoglycans. Here, we demonstrate the proof of concept of ex vivo gene editing therapy using induced pluripotent stem cell (iPSC) and CRISPR/Cas9 technologies with MPS-1 model mouse cell. Disease-affected iPSCs were generated from Idua knockout mouse embryonic fibroblasts, which carry a disrupting neomycin-resistant gene cassette (Neor) in exon VI of the Idua gene. Double guide RNAs were used to remove the Neor sequence, and various lengths of donor templates were used to reconstruct the exon VI sequence. A quantitative PCR-based screening method was used to identify Neor removal. The sequence restoration without any indel mutation was further confirmed by Sanger sequencing. After induced fibroblast differentiation, the gene-corrected iPSC-derived fibroblasts demonstrated Idua function equivalent to the wild-type iPSC-derived fibroblasts. The Idua-deficient cells were competent to be reprogrammed to iPSCs, and pluripotency was maintained through CRISPR/CAS9-mediated gene correction. These results support the concept of ex vivo gene editing therapy using iPSC and CRISPR/Cas9 technologies for MPS-1 patients.
Figures


References
-
- Pandey V. K., Tripathi A., Bhushan R., Ali A., Dubey P. K. Application of CRISPR/Cas9 genome editing in genetic disorders: a systematic review up to date. Journal of Genetic Syndromes & Gene Therapy. 2017;8(2) doi: 10.4172/2157-7412.1000321. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

