Coarse-grained dynamic RNA titration simulations
- PMID: 31065339
- PMCID: PMC6501344
- DOI: 10.1098/rsfs.2018.0066
Coarse-grained dynamic RNA titration simulations
Abstract
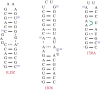
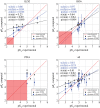
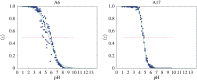
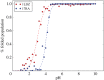
Electrostatic interactions play a pivotal role in many biomolecular processes. The molecular organization and function in biological systems are largely determined by these interactions. Owing to the highly negative charge of RNA, the effect is expected to be more pronounced in this system. Moreover, RNA base pairing is dependent on the charge of the base, giving rise to alternative secondary and tertiary structures. The equilibrium between uncharged and charged bases is regulated by the solution pH, which is therefore a key environmental condition influencing the molecule's structure and behaviour. By means of constant-pH Monte Carlo simulations based on a fast proton titration scheme, coupled with the coarse-grained model HiRE-RNA, molecular dynamic simulations of RNA molecules at constant pH enable us to explore the RNA conformational plasticity at different pH values as well as to compute electrostatic properties as local pK a values for each nucleotide.
Keywords: RNA; coarse-grained model; pH; titration.
Conflict of interest statement
We declare we have no competing interests.
Figures


References
Associated data
LinkOut - more resources
Full Text Sources
