Overexpression of mechanical sensitive miR-337-3p alleviates ectopic ossification in rat tendinopathy model via targeting IRS1 and Nox4 of tendon-derived stem cells
- PMID: 31065679
- PMCID: PMC7232128
- DOI: 10.1093/jmcb/mjz030
Overexpression of mechanical sensitive miR-337-3p alleviates ectopic ossification in rat tendinopathy model via targeting IRS1 and Nox4 of tendon-derived stem cells
Abstract
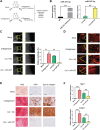
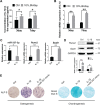
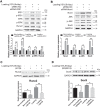
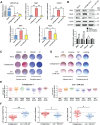
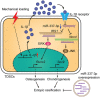
Tendinopathy, which is characterized by the ectopic ossification of tendon, is a common disease occurring in certain population, such as athletes that suffer from repetitive tendon strains. However, the molecular mechanism underlying the pathogenesis of tendinopathy caused by the overuse of tendon is still lacking. Here, we found that the mechanosensitive miRNA, miR-337-3p, had lower expression under uniaxial cyclical mechanical loading in tendon-derived stem cells (TDSCs) and negatively controlled chondro-osteogenic differentiation of TDSCs. Importantly, downregulation of miR-337-3p expression was also observed in both rat and human calcified tendons, and overexpressing miR-337-3p in patellar tendons of rat tendinopathy model displayed a robust therapeutic efficiency. Mechanistically, we found that the proinflammatory cytokine interleukin-1β was the upstream factor of miR-337-3p that bridges the mechanical loading with its downregulation. Furthermore, the target genes of miR-337-3p, NADPH oxidase 4, and insulin receptor substrate 1, activated chondro-osteogenic differentiation of TDSCs through JNK and ERK signaling, respectively. Thus, these findings not only provide novel insight into the molecular mechanisms underlying ectopic ossification in tendinopathy but also highlight the significance of miR-337-3p as a putative therapeutic target for clinic treatment of tendinopathy.
Keywords: chondro-osteogenesis; mechanical loading; mechanosensitive miRNA; tendinopathy; tendon-derived stem cells.
© The Author(s) (2019). Published by Oxford University Press on behalf of Journal of Molecular Cell Biology, IBCB, SIBS, CAS.
Figures


References
-
- Ambros V. (2004). The functions of animal microRNAs. Nature 431, 350–355. - PubMed
-
- Aspenberg P. (2007). Is inflammation harmless to loaded tendons? J. Appl. Physiol. 102, 3–4. - PubMed
-
- Barwari T., Joshi A., and Mayr M. (2016). MicroRNAs in cardiovascular disease. J. Am. Coll. Cardiol. 68, 2577–2584. - PubMed
-
- Berkoff D.J., Kallianos S.A., Eskildsen S.M., et al. (2016). Use of an IL1-receptor antagonist to prevent the progression of tendinopathy in a rat model. J. Orthop. Res. 34, 616–622. - PubMed
-
- Bi Y., Ehirchiou D., Kilts T.M., et al. (2007). Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat. Med. 13, 1219–1227. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

