Natural Language Processing of Clinical Notes on Chronic Diseases: Systematic Review
- PMID: 31066697
- PMCID: PMC6528438
- DOI: 10.2196/12239
Natural Language Processing of Clinical Notes on Chronic Diseases: Systematic Review
Abstract
Background: Novel approaches that complement and go beyond evidence-based medicine are required in the domain of chronic diseases, given the growing incidence of such conditions on the worldwide population. A promising avenue is the secondary use of electronic health records (EHRs), where patient data are analyzed to conduct clinical and translational research. Methods based on machine learning to process EHRs are resulting in improved understanding of patient clinical trajectories and chronic disease risk prediction, creating a unique opportunity to derive previously unknown clinical insights. However, a wealth of clinical histories remains locked behind clinical narratives in free-form text. Consequently, unlocking the full potential of EHR data is contingent on the development of natural language processing (NLP) methods to automatically transform clinical text into structured clinical data that can guide clinical decisions and potentially delay or prevent disease onset.
Objective: The goal of the research was to provide a comprehensive overview of the development and uptake of NLP methods applied to free-text clinical notes related to chronic diseases, including the investigation of challenges faced by NLP methodologies in understanding clinical narratives.
Methods: Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines were followed and searches were conducted in 5 databases using "clinical notes," "natural language processing," and "chronic disease" and their variations as keywords to maximize coverage of the articles.
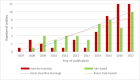
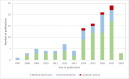
Results: Of the 2652 articles considered, 106 met the inclusion criteria. Review of the included papers resulted in identification of 43 chronic diseases, which were then further classified into 10 disease categories using the International Classification of Diseases, 10th Revision. The majority of studies focused on diseases of the circulatory system (n=38) while endocrine and metabolic diseases were fewest (n=14). This was due to the structure of clinical records related to metabolic diseases, which typically contain much more structured data, compared with medical records for diseases of the circulatory system, which focus more on unstructured data and consequently have seen a stronger focus of NLP. The review has shown that there is a significant increase in the use of machine learning methods compared to rule-based approaches; however, deep learning methods remain emergent (n=3). Consequently, the majority of works focus on classification of disease phenotype with only a handful of papers addressing extraction of comorbidities from the free text or integration of clinical notes with structured data. There is a notable use of relatively simple methods, such as shallow classifiers (or combination with rule-based methods), due to the interpretability of predictions, which still represents a significant issue for more complex methods. Finally, scarcity of publicly available data may also have contributed to insufficient development of more advanced methods, such as extraction of word embeddings from clinical notes.
Conclusions: Efforts are still required to improve (1) progression of clinical NLP methods from extraction toward understanding; (2) recognition of relations among entities rather than entities in isolation; (3) temporal extraction to understand past, current, and future clinical events; (4) exploitation of alternative sources of clinical knowledge; and (5) availability of large-scale, de-identified clinical corpora.
Keywords: cancer; chronic diseases; clinical notes; deep learning; diabetes; electronic health records; heart disease; lung disease; machine learning; natural language processing; stroke.
©Seyedmostafa Sheikhalishahi, Riccardo Miotto, Joel T Dudley, Alberto Lavelli, Fabio Rinaldi, Venet Osmani. Originally published in JMIR Medical Informatics (http://medinform.jmir.org), 27.04.2019.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures


References
-
- World Health Organization. [2019-03-29]. WHO Global status report on noncommunicable diseases 2014 https://www.who.int/nmh/publications/ncd-status-report-2014/en/
-
- Miotto R, Li L, Kidd BA, Dudley JT. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep. 2016 Dec 17;6:26094. doi: 10.1038/srep26094. http://paperpile.com/b/UsJJXQ/OwN0t srep26094 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

