ClpP Protease, a Promising Antimicrobial Target
- PMID: 31067645
- PMCID: PMC6540193
- DOI: 10.3390/ijms20092232
ClpP Protease, a Promising Antimicrobial Target
Abstract
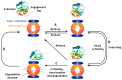
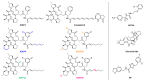
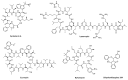
The caseinolytic protease proteolytic subunit (ClpP) is a serine protease playing an important role in proteostasis of eukaryotic organelles and prokaryotic cells. Alteration of ClpP function has been proved to affect the virulence and infectivity of a number of pathogens. Increased bacterial resistance to antibiotics has become a global problem and new classes of antibiotics with novel mechanisms of action are needed. In this regard, ClpP has emerged as an attractive and potentially viable option to tackle pathogen fitness without suffering cross-resistance to established antibiotic classes and, when not an essential target, without causing an evolutionary selection pressure. This opens a greater window of opportunity for the host immune system to clear the infection by itself or by co-administration with commonly prescribed antibiotics. A comprehensive overview of the function, regulation and structure of ClpP across the different organisms is given. Discussion about mechanism of action of this protease in bacterial pathogenesis and human diseases are outlined, focusing on the compounds developed in order to target the activation or inhibition of ClpP.
Keywords: ClpP; antibiotics; antivirulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
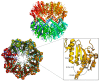


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

