eCD4-Ig Limits HIV-1 Escape More Effectively than CD4-Ig or a Broadly Neutralizing Antibody
- PMID: 31068428
- PMCID: PMC6600210
- DOI: 10.1128/JVI.00443-19
eCD4-Ig Limits HIV-1 Escape More Effectively than CD4-Ig or a Broadly Neutralizing Antibody
Abstract
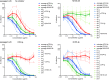
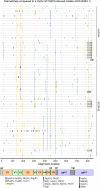
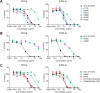
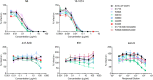
The engineered antibody-like entry inhibitor eCD4-Ig neutralizes every human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus isolate it has been tested against. The exceptional breadth of eCD4-Ig derives from its ability to closely and simultaneously emulate the HIV-1 receptor CD4 and coreceptors, either CCR5 or CXCR4. Here we investigated whether viral escape from eCD4-Ig is more difficult than that from CD4-Ig or the CD4-binding site antibody NIH45-46. We observed that a viral swarm selected with high concentrations of eCD4-Ig was increasingly resistant to but did not fully escape from eCD4-Ig. In contrast, viruses selected under the same conditions with CD4-Ig or NIH45-46 fully escaped from those inhibitors. eCD4-Ig-resistant viruses acquired unique changes in the V2 apex, V3, V4, and CD4-binding regions of the HIV-1 envelope glycoprotein (Env). Most of the alterations did not directly affect neutralization by eCD4-Ig or neutralizing antibodies. However, alteration of Q428 to an arginine or lysine resulted in markedly greater resistance to eCD4-Ig and CD4-Ig, with correspondingly dramatic losses in infectivity and greater sensitivity to a V3 antibody and to serum from an infected individual. Compensatory mutations in the V3 loop (N301D) and in the V2 apex (K171E) partially restored viral fitness without affecting serum or eCD4-Ig sensitivity. Collectively, these data suggest that multiple mutations will be necessary to fully escape eCD4-Ig without loss of viral fitness.IMPORTANCE HIV-1 broadly neutralizing antibodies (bNAbs) and engineered antibody-like inhibitors have been compared for their breadths, potencies, and in vivo half-lives. However, a key limitation in the use of antibodies to treat an established HIV-1 infection is the rapid emergence of fully resistant viruses. Entry inhibitors of similar breadths and potencies can differ in the ease with which viral escape variants arise. Here we show that HIV-1 escape from the potent and exceptionally broad entry inhibitor eCD4-Ig is more difficult than that from CD4-Ig or the bNAb NIH45-46. Indeed, full escape was not observed under conditions under which escape from CD4-Ig or NIH45-46 was readily detected. Moreover, viruses that were partially resistant to eCD4-Ig were markedly less infective and more sensitive to antibodies in the serum of an infected person. These data suggest that eCD4-Ig will be more difficult to escape and that even partial escape will likely extract a high fitness cost.
Keywords: CCR5; CD4; HIV-1; eCD4-Ig; viral entry.
Copyright © 2019 American Society for Microbiology.
Figures



Similar articles
-
eCD4-Ig Variants That More Potently Neutralize HIV-1.J Virol. 2018 May 29;92(12):e02011-17. doi: 10.1128/JVI.02011-17. Print 2018 Jun 15. J Virol. 2018. PMID: 29593050 Free PMC article.
-
A Coreceptor-Mimetic Peptide Enhances the Potency of V3-Glycan Antibodies.J Virol. 2019 Feb 19;93(5):e01653-18. doi: 10.1128/JVI.01653-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30541842 Free PMC article.
-
AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges.Nature. 2015 Mar 5;519(7541):87-91. doi: 10.1038/nature14264. Epub 2015 Feb 18. Nature. 2015. PMID: 25707797 Free PMC article.
-
Conformation-Dependent Interactions Between HIV-1 Envelope Glycoproteins and Broadly Neutralizing Antibodies.AIDS Res Hum Retroviruses. 2018 Sep;34(9):794-803. doi: 10.1089/AID.2018.0102. Epub 2018 Jul 17. AIDS Res Hum Retroviruses. 2018. PMID: 29905080 Review.
-
Engineering multi-specific antibodies against HIV-1.Retrovirology. 2018 Aug 29;15(1):60. doi: 10.1186/s12977-018-0439-9. Retrovirology. 2018. PMID: 30157871 Free PMC article. Review.
Cited by
-
Asymmetric opening of HIV-1 Env bound to CD4 and a coreceptor-mimicking antibody.Nat Struct Mol Biol. 2019 Dec;26(12):1167-1175. doi: 10.1038/s41594-019-0344-5. Epub 2019 Dec 2. Nat Struct Mol Biol. 2019. PMID: 31792452 Free PMC article.
-
The Conformational States of the HIV-1 Envelope Glycoproteins.Trends Microbiol. 2020 Aug;28(8):655-667. doi: 10.1016/j.tim.2020.03.007. Epub 2020 May 14. Trends Microbiol. 2020. PMID: 32418859 Free PMC article. Review.
-
In vivo evolution of env in SHIV-AD8EO-infected rhesus macaques after AAV-vectored delivery of eCD4-Ig.Mol Ther. 2025 Feb 5;33(2):560-579. doi: 10.1016/j.ymthe.2024.12.015. Epub 2024 Dec 12. Mol Ther. 2025. PMID: 39673132 Free PMC article.
-
Nanoparticles presenting clusters of CD4 expose a universal vulnerability of HIV-1 by mimicking target cells.Proc Natl Acad Sci U S A. 2020 Aug 4;117(31):18719-18728. doi: 10.1073/pnas.2010320117. Epub 2020 Jul 20. Proc Natl Acad Sci U S A. 2020. PMID: 32690692 Free PMC article.
-
Deep Mutational Scanning of Viral Glycoproteins and Their Host Receptors.Front Mol Biosci. 2021 Apr 9;8:636660. doi: 10.3389/fmolb.2021.636660. eCollection 2021. Front Mol Biosci. 2021. PMID: 33898517 Free PMC article. Review.
References
-
- Liao H-X, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, NISC Comparative Sequencing Program, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia S-M, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BTM, Kwong PD, Mascola JR, Haynes BF. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496:469–476. doi:10.1038/nature12053. - DOI - PMC - PubMed
-
- Bonsignori M, Zhou T, Sheng Z, Chen L, Gao F, Joyce MG, Ozorowski G, Chuang G-Y, Schramm CA, Wiehe K, Alam SM, Bradley T, Gladden MA, Hwang K-K, Iyengar S, Kumar A, Lu X, Luo K, Mangiapani MC, Parks RJ, Song H, Acharya P, Bailer RT, Cao A, Druz A, Georgiev IS, Kwon YD, Louder MK, Zhang B, Zheng A, Hill BJ, Kong R, Soto C, Mullikin JC, Douek DC, Montefiori DC, Moody MA, Shaw GM, Hahn BH, Kelsoe G, Hraber PT, Korber BT, Boyd SD, Fire AZ, Kepler TB, Shapiro L, Ward AB, Mascola JR, Liao H-X, Kwong PD, Haynes BF. 2016. Maturation pathway from germline to broad HIV-1 neutralizer of a CD4-mimic antibody. Cell 165:449–463. doi:10.1016/j.cell.2016.02.022. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

