Mucosal-associated invariant T cells and oral microbiome in persistent apical periodontitis
- PMID: 31068577
- PMCID: PMC6506549
- DOI: 10.1038/s41368-019-0049-y
Mucosal-associated invariant T cells and oral microbiome in persistent apical periodontitis
Abstract
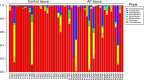
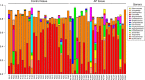
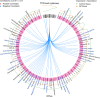
Opportunistic bacteria in apical periodontitis (AP) may pose a risk for systemic dissemination. Mucosal-associated invariant T (MAIT) cells are innate-like T cells with a broad and potent antimicrobial activity important for gut mucosal integrity. It was recently shown that MAIT cells are present in the oral mucosal tissue, but the involvement of MAIT cells in AP is unknown. Here, comparison of surgically resected AP and gingival tissues demonstrated that AP tissues express significantly higher levels of Vα7.2-Jα33, Vα7.2-Jα20, Vα7.2-Jα12, Cα and tumour necrosis factor (TNF), interferon (IFN)-γ and interleukin (IL)-17A transcripts, resembling a MAIT cell signature. Moreover, in AP tissues the MR1-restricted MAIT cells positive for MR1-5-OP-RU tetramer staining appeared to be of similar levels as in peripheral blood but consisted mainly of CD4+ subset. Unlike gingival tissues, the AP microbiome was quantitatively impacted by factors like fistula and high patient age and had a prominent riboflavin-expressing bacterial feature. When merged in an integrated view, the examined immune and microbiome data in the sparse partial least squares discriminant analysis could identify bacterial relative abundances that negatively correlated with Vα7.2-Jα33, Cα, and IL-17A transcript expressions in AP, implying that MAIT cells could play a role in the local defence at the oral tissue barrier. In conclusion, we describe the presence of MAIT cells at the oral site where translocation of oral microbiota could take place. These findings have implications for understanding the immune sensing of polymicrobial-related oral diseases.
Conflict of interest statement
The authors declare no competing interests.
Figures








References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

