A longitudinal big data approach for precision health
- PMID: 31068711
- PMCID: PMC6713274
- DOI: 10.1038/s41591-019-0414-6
A longitudinal big data approach for precision health
Abstract
Precision health relies on the ability to assess disease risk at an individual level, detect early preclinical conditions and initiate preventive strategies. Recent technological advances in omics and wearable monitoring enable deep molecular and physiological profiling and may provide important tools for precision health. We explored the ability of deep longitudinal profiling to make health-related discoveries, identify clinically relevant molecular pathways and affect behavior in a prospective longitudinal cohort (n = 109) enriched for risk of type 2 diabetes mellitus. The cohort underwent integrative personalized omics profiling from samples collected quarterly for up to 8 years (median, 2.8 years) using clinical measures and emerging technologies including genome, immunome, transcriptome, proteome, metabolome, microbiome and wearable monitoring. We discovered more than 67 clinically actionable health discoveries and identified multiple molecular pathways associated with metabolic, cardiovascular and oncologic pathophysiology. We developed prediction models for insulin resistance by using omics measurements, illustrating their potential to replace burdensome tests. Finally, study participation led the majority of participants to implement diet and exercise changes. Altogether, we conclude that deep longitudinal profiling can lead to actionable health discoveries and provide relevant information for precision health.
Figures

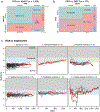




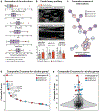



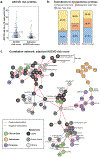
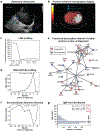

Comment in
-
The future of individualized health maintenance.Nat Med. 2019 May;25(5):712-714. doi: 10.1038/s41591-019-0443-1. Nat Med. 2019. PMID: 31068704 No abstract available.
References
-
- National Research Council (US) Committee on A Framework for Developing a New Taxonomy of Disease. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. (National Academies Press (US), 2012). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

