Identifying Gut Microbiota Associated With Colorectal Cancer Using a Zero-Inflated Lognormal Model
- PMID: 31068913
- PMCID: PMC6491826
- DOI: 10.3389/fmicb.2019.00826
Identifying Gut Microbiota Associated With Colorectal Cancer Using a Zero-Inflated Lognormal Model
Abstract
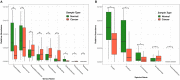
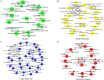
Colorectal cancer (CRC) is the third most common cancer worldwide. Its incidence is still increasing, and the mortality rate is high. New therapeutic and prognostic strategies are urgently needed. It became increasingly recognized that the gut microbiota composition differs significantly between healthy people and CRC patients. Thus, identifying the difference between gut microbiota of the healthy people and CRC patients is fundamental to understand these microbes' functional roles in the development of CRC. We studied the microbial community structure of a CRC metagenomic dataset of 156 patients and healthy controls, and analyzed the diversity, differentially abundant bacteria, and co-occurrence networks. We applied a modified zero-inflated lognormal (ZIL) model for estimating the relative abundance. We found that the abundance of genera: Anaerostipes, Bilophila, Catenibacterium, Coprococcus, Desulfovibrio, Flavonifractor, Porphyromonas, Pseudoflavonifractor, and Weissella was significantly different between the healthy and CRC groups. We also found that bacteria such as Streptococcus, Parvimonas, Collinsella, and Citrobacter were uniquely co-occurring within the CRC patients. In addition, we found that the microbial diversity of healthy controls is significantly higher than that of the CRC patients, which indicated a significant negative correlation between gut microbiota diversity and the stage of CRC. Collectively, our results strengthened the view that individual microbes as well as the overall structure of gut microbiota were co-evolving with CRC.
Keywords: association network; colorectal cancer; gut microbiota; microbial diversity; zero-inflated lognormal model.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

