Positive Selection Evidence in Xylose-Related Genes Suggests Methylglyoxal Reductase as a Target for the Improvement of Yeasts' Fermentation in Industry
- PMID: 31070742
- PMCID: PMC6637916
- DOI: 10.1093/gbe/evz036
Positive Selection Evidence in Xylose-Related Genes Suggests Methylglyoxal Reductase as a Target for the Improvement of Yeasts' Fermentation in Industry
Abstract
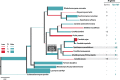
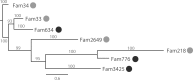
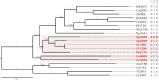
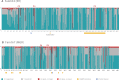
Xylose assimilation and fermentation are important traits for second generation ethanol production. However, some genomic features associated with this pentose sugar's metabolism remain unknown in yeasts. Comparative genomics studies have led to important insights in this field, but we are still far from completely understanding endogenous yeasts' xylose metabolism. In this work, we carried out a deep evolutionary analysis suited for comparative genomics of xylose-consuming yeasts, searching for of positive selection on genes associated with glucose and xylose metabolism in the xylose-fermenters' clade. Our investigation detected positive selection fingerprints at this clade not only among sequences of important genes for xylose metabolism, such as xylose reductase and xylitol dehydrogenase, but also in genes expected to undergo neutral evolution, such as the glycolytic gene phosphoglycerate mutase. In addition, we present expansion, positive selection marks, and convergence as evidence supporting the hypothesis that natural selection is shaping the evolution of the little studied methylglyoxal reductases. We propose a metabolic model suggesting that selected codons among these proteins caused a putative change in cofactor preference from NADPH to NADH that alleviates cellular redox imbalance. These findings provide a wider look into pentose metabolism of yeasts and add this previously overlooked piece into the intricate puzzle of oxidative imbalance. Although being extensively discussed in evolutionary works the awareness of selection patterns is recent in biotechnology researches, rendering insights to surpass the reached status quo in many of its subareas.
Keywords: Saccharomycotina; comparative genomics; natural selection; phylogenetics; xylitol dehydrogenase; xylose reductase.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Aditiya HB, Mahlia TMI, Chong WT, Nur H, Sebayang AH.. 2016. Second generation bioethanol production: a critical review. Renew Sustain Energy Rev. 66:631–653.
-
- Bajwa PK, Harrington S, Dashtban M, Lee H.. 2016. Expression and characterization of glycosyl hydrolase family 115 α-glucuronidase from Scheffersomyces stipitis. Ind Biotechnol. 12(2):98–104.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

