N6-methyladenosine regulatory machinery in plants: composition, function and evolution
- PMID: 31070865
- PMCID: PMC6576107
- DOI: 10.1111/pbi.13149
N6-methyladenosine regulatory machinery in plants: composition, function and evolution
Abstract
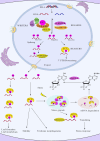
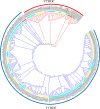
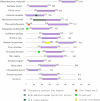
N6-methyladenosine (m6A) RNA methylation, one of the most pivotal internal modifications of RNA, is a conserved post-transcriptional mechanism to enrich and regulate genetic information in eukaryotes. The scope and function of this modification in plants has been an intense focus of study, especially in model plant systems. The characterization of plant m6A writers, erasers and readers, as well as the elucidation of their functions, is currently one of the most fascinating hotspots in plant biology research. The functional analysis of m6A in plants will be booming in the foreseeable future, which could contribute to crop genetic improvement through epitranscriptome manipulation. In this review, we systematically analysed and summarized recent advances in the understanding of the structure and composition of plant m6A regulatory machinery, and the biological functions of m6A in plant growth, development and stress response. Finally, our analysis showed that the evolutionary relationships between m6A modification components were highly conserved across the plant kingdom.
Keywords: RNA modification; epitranscriptome; m6A; plant; regulatory machinery.
© 2019 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Arguello, A.E. , DeLiberto, A.N. and Kleiner, R.E. (2017) RNA chemical proteomics reveals the N6‐methyladenosine (m6A)‐regulated protein–RNA interactome. J. Am. Chem. Soc. 139, 17249–17252. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

