Bacteroides-Derived Sphingolipids Are Critical for Maintaining Intestinal Homeostasis and Symbiosis
- PMID: 31071294
- PMCID: PMC6544385
- DOI: 10.1016/j.chom.2019.04.002
Bacteroides-Derived Sphingolipids Are Critical for Maintaining Intestinal Homeostasis and Symbiosis
Abstract
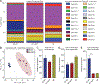
Sphingolipids are structural membrane components and important eukaryotic signaling molecules. Sphingolipids regulate inflammation and immunity and were recently identified as the most differentially abundant metabolite in stool from inflammatory bowel disease (IBD) patients. Commensal bacteria from the Bacteroidetes phylum also produce sphingolipids, but the impact of these metabolites on host pathways is largely uncharacterized. To determine whether bacterial sphingolipids modulate intestinal health, we colonized germ-free mice with a sphingolipid-deficient Bacteroides thetaiotaomicron strain. A lack of Bacteroides-derived sphingolipids resulted in intestinal inflammation and altered host ceramide pools in mice. Using lipidomic analysis, we described a sphingolipid biosynthesis pathway and revealed a variety of Bacteroides-derived sphingolipids including ceramide phosphoinositol and deoxy-sphingolipids. Annotating Bacteroides sphingolipids in an IBD metabolomic dataset revealed lower abundances in IBD and negative correlations with inflammation and host sphingolipid production. These data highlight the role of bacterial sphingolipids in maintaining homeostasis and symbiosis in the gut.
Keywords: Bacteroides; inflammation; inflammatory bowel disease; innate immunity; metabolism; microbiome; sphingolipids.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures
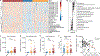



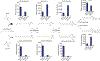

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

