A Type III CRISPR Ancillary Ribonuclease Degrades Its Cyclic Oligoadenylate Activator
- PMID: 31071326
- PMCID: PMC6599890
- DOI: 10.1016/j.jmb.2019.04.041
A Type III CRISPR Ancillary Ribonuclease Degrades Its Cyclic Oligoadenylate Activator
Abstract
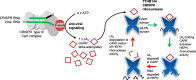
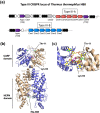
Cyclic oligoadenylate (cOA) secondary messengers are generated by type III CRISPR systems in response to viral infection. cOA allosterically activates the CRISPR ancillary ribonucleases Csx1/Csm6, which degrade RNA non-specifically using a HEPN (Higher Eukaryotes and Prokaryotes, Nucleotide binding) active site. This provides effective immunity but can also lead to growth arrest in infected cells, necessitating a means to deactivate the ribonuclease once viral infection has been cleared. In the crenarchaea, dedicated ring nucleases degrade cA4 (cOA consisting of 4 AMP units), but the equivalent enzyme has not been identified in bacteria. We demonstrate that, in Thermus thermophilus HB8, the uncharacterized protein TTHB144 is a cA4-activated HEPN ribonuclease that also degrades its activator. TTHB144 binds and degrades cA4 at an N-terminal CARF (CRISPR-associated Rossman fold) domain. The two activities can be separated by site-directed mutagenesis. TTHB144 is thus the first example of a self-limiting CRISPR ribonuclease.
Keywords: CRISPR; anti-viral signaling; cyclic oligoadenylate; ring nuclease, Thermus thermophilus.
Copyright © 2019 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Figures


References
-
- Koonin E.V., Makarova K.S. Mobile genetic elements and evolution of CRISPR–Cas systems: all the way there and back. Genome Biol. Evol. 2017;9:2812–2825. - PMC - PubMed
- E. V. Koonin, K.S. Makarova, Mobile Genetic Elements and Evolution of CRISPR-Cas Systems: All the Way There and Back, Genome Biol. Evol. 9 (2017) 2812–2825. - PMC - PubMed
-
- Hille F., Richter H., Wong S.P., Bratovič M., Sarah R., Charpentier E. The biology of CRISPR–Cas: backward and forward. Cell. 2018;172:1239–1259. - PubMed
- F. Hille, H. Richter, S.P. Wong, M. Bratovič, R. Sarah, E. Charpentier, The Biology of CRISPR-Cas: Backward and Forward, Cell. 172 (2018) 1239–1259. - PubMed
-
- G. Tamulaitis, Č. Venclovas, V. Siksnys, Type III CRISPR–Cas immunity: major differences brushed aside, Trends Microbiol. 25 (2017) 49–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

