Effect of Drought Stress and Developmental Stages on Microbial Community Structure and Diversity in Peanut Rhizosphere Soil
- PMID: 31071918
- PMCID: PMC6540327
- DOI: 10.3390/ijms20092265
Effect of Drought Stress and Developmental Stages on Microbial Community Structure and Diversity in Peanut Rhizosphere Soil
Abstract
Background: Peanut (Arachis hypogaea L.), an important oilseed and food legume, is widely cultivated in the semi-arid tropics. Drought is the major stress in this region which limits productivity. Microbial communities in the rhizosphere are of special importance to stress tolerance. However, relatively little is known about the relationship between drought and microbial communities in peanuts.
Method: In this study, deep sequencing of the V3-V4 region of the 16S rRNA gene was performed to characterize the microbial community structure of drought-treated and untreated peanuts.
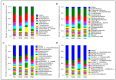
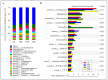
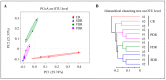
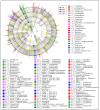
Results: Taxonomic analysis showed that Actinobacteria, Proteobacteria, Saccharibacteria, Chloroflexi, Acidobacteria and Cyanobacteria were the dominant phyla in the peanut rhizosphere. Comparisons of microbial community structure of peanuts revealed that the relative abundance of Actinobacteria and Acidobacteria dramatically increased in the seedling and podding stages in drought-treated soil, while that of Cyanobacteria and Gemmatimonadetes increased in the flowering stage in drought-treated rhizospheres. Metagenomic profiling indicated that sequences related to metabolism, signaling transduction, defense mechanism and basic vital activity were enriched in the drought-treated rhizosphere, which may have implications for plant survival and drought tolerance.
Conclusion: This microbial communities study will form the foundation for future improvement of drought tolerance of peanuts via modification of the soil microbes.
Keywords: drought stress; microbial community structure; peanut (Arachis hypogaea L.); rhizosphere.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Cuc L.M., Mace E.S., Crouch J.H., Quang V.D., Long T.D., Varshney R.K. Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea) BMC Plant Biol. 2008;8:55. doi: 10.1186/1471-2229-8-55. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

