Infection and nuclear interaction in mammalian cells by 'Candidatus Berkiella cookevillensis', a novel bacterium isolated from amoebae
- PMID: 31072343
- PMCID: PMC6507137
- DOI: 10.1186/s12866-019-1457-z
Infection and nuclear interaction in mammalian cells by 'Candidatus Berkiella cookevillensis', a novel bacterium isolated from amoebae
Abstract
Background: 'Candidatus Berkiella cookevillensis' and 'Ca. Berkiella aquae' have previously been described as intranuclear bacteria of amoebae. Both bacteria were isolated from amoebae and were described as appearing within the nuclei of Acanthamoeba polyphaga and ultimately lysing their host cells within 4 days. Both bacteria are Gammaproteobacteria in the order Legionellales with the greatest similarity to Coxiella burnetii. Neither bacterium grows axenically in artificial culture media. In this study, we further characterized 'Ca. B. cookevillensis' by demonstrating association with nuclei of human phagocytic and nonphagocytic cell lines.
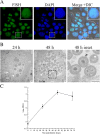
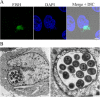
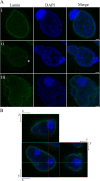
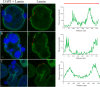
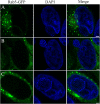
Results: Transmission electron microscopy (TEM) and confocal microscopy were used to confirm nuclear co-localization of 'Ca. B. cookevillensis' in the amoeba host A. polyphaga with 100% of cells having bacteria co-localized with host nuclei by 48 h. TEM and confocal microscopy demonstrated that the bacterium was also observed to be closely associated with nuclei of human U937 and THP-1 differentiated macrophage cell lines and nonphagocytic HeLa human epithelial-like cells. Immunofluorescent staining revealed that the bacteria-containing vacuole invaginates the nuclear membranes and appears to cross from the cytoplasm into the nucleus as an intact vacuole.
Conclusion: Results of this study indicate that a novel coccoid bacterium isolated from amoebae can infect human cell lines by associating with the host cell nuclei, either by crossing the nuclear membranes or by deeply invaginating the nuclear membranes. When associated with the nuclei, the bacteria appear to be bound within a vacuole and replicate to high numbers by 48 h. We believe this is the first report of such a process involving bacteria and human cell lines.
Keywords: Bacteria; Coxiella; Endosymbiont; Human; Legionella; Nucleus; Symbiosis.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Mehari YT, Hayes BJ, Redding KS, Mariappan PV, Gunderson JH, Farone AL, Farone MB. Description of ‘Candidatus Berkiella aquae’ and ‘Candidatus Berkiella cookevillensis’, two intranuclear bacteria of freshwater amoebae. Int J Syst Evol Microbiol. 2016;66:536–541. doi: 10.1099/ijsem.0.000750. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

