Transcriptome and physiological analyses for revealing genes involved in wheat response to endoplasmic reticulum stress
- PMID: 31072347
- PMCID: PMC6509841
- DOI: 10.1186/s12870-019-1798-7
Transcriptome and physiological analyses for revealing genes involved in wheat response to endoplasmic reticulum stress
Abstract
Background: Wheat production is largely restricted by adverse environmental stresses. Under many undesirable conditions, endoplasmic reticulum (ER) stress can be induced. However, the physiological and molecular responses of wheat to ER stress remain poorly understood. We used dithiothreitol (DTT) and tauroursodeoxycholic acid (TUDCA) to induce or suppress ER stress in wheat cells, respectively, with the aim to reveal the molecular background of ER stress responses using a combined approach of transcriptional profiling and morpho-physiological characterization.
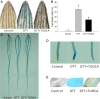
Methods: To understand the mechanism of wheat response to ER stress, three wheat cultivars were used in our pre-experiments. Among them, the cultivar with a moderate stress tolerance, Yunong211 was used in the following experiments. We used DTT (7.5 mM) to induce ER stress and TUDCA (25 μg·mL- 1) to suppress the stress. Under three treatment groups (Control, DTT and DTT + TUDCA), we firstly monitored the morphological, physiological and cytological changes of wheat seedlings. Then we collected leaf samples from each group for RNA extraction, library construction and RNA sequencing on an Illumina Hiseq platform. The sequencing data was then validated by qRT-PCR.
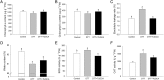
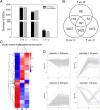
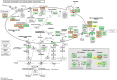
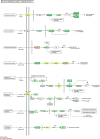
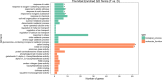
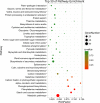
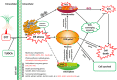
Results: Morpho-physiological results showed DTT significantly reduced plant height and biomass, decreased contents of chlorophyll and water, increased electrolyte leakage rate and antioxidant enzymes activity, and accelerated the cell death ratio, whereas these changes were all remarkably alleviated after TUDCA co-treatment. Therefore, RNA sequencing was performed to determine the genes involved in regulating wheat response to stress. Transcriptomic analysis revealed that 8204 genes were differentially expressed in three treatment groups. Among these genes, 158 photosynthesis-related genes, 42 antioxidant enzyme genes, 318 plant hormone-related genes and 457 transcription factors (TFs) may play vital roles in regulating wheat response to ER stress. Based on the comprehensive analysis, we propose a hypothetical model to elucidate possible mechanisms of how plants adapt to environmental stresses.
Conclusions: We identified several important genes that may play vital roles in wheat responding to ER stress. This work should lay the foundations of future studies in plant response to environmental stresses.
Keywords: Antioxidant enzymes; Chlorophyll; DTT; RNA-seq; TUDCA; Transcription factors.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

