Assessment of total, ligand-induced peroxisome proliferator activated receptor γ ligand activity in serum
- PMID: 31072366
- PMCID: PMC6506953
- DOI: 10.1186/s12940-019-0486-2
Assessment of total, ligand-induced peroxisome proliferator activated receptor γ ligand activity in serum
Abstract
Background: Humans are exposed to a complex mixture of environmental chemicals that impact bone and metabolic health, and traditional exposure assessments struggle to capture these exposure scenarios. Peroxisome proliferator activated receptor-gamma (PPARγ) is an essential regulator of metabolic and bone homeostasis, and its inappropriate activation by environmental chemicals can set the stage for adverse health effects. Here, we present the development of the Serum PPARγ Activity Assay (SPAA), a simple and cost-effective method to measure total ligand activity in small volumes of serum.
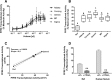
Methods: First, we determined essential components of the bioassay. Cos-7 cells were transfected with combinations of expression vectors for human PPARγ and RXRα, the obligate DNA-binding partner of PPARγ, along with PPRE (DR1)-driven luciferase and control eGFP reporter constructs. Transfected cells were treated with rosiglitazone, a synthetic PPARγ ligand and/or LG100268, a synthetic RXR ligand, to characterize the dose response and determine the simplest and most efficacious format. Following optimization of the bioassay, we assessed the cumulative activation of PPARγ by ligands in serum from mice treated with a PPARγ ligand and commercial human serum samples.
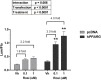
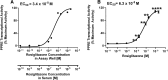
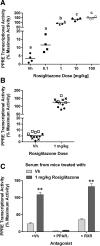
Results: Cos-7 cells endogenously express sufficient RXR to support efficacious activation of transfected PPARγ. Co-transfection of an RXR expression vector with the PPARγ expression vector did not increase PPRE transcriptional activity induced by rosiglitazone. Treatment with an RXR ligand marginally increased PPRE transcriptional activity in the presence of transfected PPARγ, and co-treatment with an RXR ligand reduced rosiglitazone-induced PPRE transcriptional activity. Therefore, the final bioassay protocol consists of transfecting Cos-7 cells with a PPARγ expression vector along with the reporter vectors, applying rosiglitazone standards and/or 10 μL of serum, and measuring luminescence and fluorescence after a 24 h incubation. Sera from mice dosed with rosiglitazone induced PPRE transcriptional activity in the SPAA in a dose-dependent and PPARγ-dependent manner. Additionally, human serum from commercial sources induced a range of PPRE transcriptional activities in a PPARγ-dependent manner, demonstrating the ability of the bioassay to detect potentially low levels of ligands.
Conclusions: The SPAA can reliably measure total PPRE transcriptional activity in small volumes of serum. This system provides a sensitive, straightforward assay that can be reproduced in any cell culture laboratory.
Keywords: Human serum; Metabolism disrupting compounds; Mixtures; PPARγ.
Conflict of interest statement
Ethics approval and consent to participate
Animal studies were reviewed and approved by the Institutional Animal Care and Use Committee at Boston University and performed in an American Association for the Accreditation of Laboratory Animal Care accredited facility (Animal Welfare Assurance Number: A3316–01). All animals were treated humanely and with regard for alleviation of suffering.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



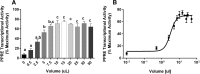

References
-
- Pool R, Rusch E, Institute of Medicine (U.S.). roundtable on environmental health sciences research and medicine: identifying and reducing environmental health risks of chemicals in our society : workshop summary. Washington, D.C.: the National Academies Press; 2014. - PubMed
-
- Myth versus fact about chemicals in commerce. [http://www.socma.com/advocacy/issues/chemical-risk-management/myth-versu...].
-
- Heindel Jerrold J., Blumberg Bruce, Cave Mathew, Machtinger Ronit, Mantovani Alberto, Mendez Michelle A., Nadal Angel, Palanza Paola, Panzica Giancarlo, Sargis Robert, Vandenberg Laura N., vom Saal Frederick. Metabolism disrupting chemicals and metabolic disorders. Reproductive Toxicology. 2017;68:3–33. - PMC - PubMed
-
- Anghel SI, Bedu E, Vivier CD, Descombes P, Desvergne B, Wahli W. Adipose tissue integrity as a prerequisite for systemic energy balance: a critical role for peroxisome proliferator-activated receptor gamma. J Biol Chem. 2007;282(41):29946–29957. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

