Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes
- PMID: 31073061
- PMCID: PMC7034192
- DOI: 10.1126/science.aaw5188
Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes
Abstract
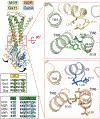
Muscarinic acetylcholine receptors are G protein-coupled receptors that respond to acetylcholine and play important signaling roles in the nervous system. There are five muscarinic receptor subtypes (M1R to M5R), which, despite sharing a high degree of sequence identity in the transmembrane region, couple to different heterotrimeric GTP-binding proteins (G proteins) to transmit signals. M1R, M3R, and M5R couple to the Gq/ 11 family, whereas M2R and M4R couple to the Gi/ o family. Here, we present and compare the cryo-electron microscopy structures of M1R in complex with G11 and M2R in complex with GoA The M1R-G11 complex exhibits distinct features, including an extended transmembrane helix 5 and carboxyl-terminal receptor tail that interacts with G protein. Detailed analysis of these structures provides a framework for understanding the molecular determinants of G-protein coupling selectivity.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures

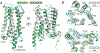

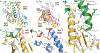
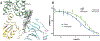
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

