Epigenome-wide association study of lung function level and its change
- PMID: 31073081
- PMCID: PMC6610463
- DOI: 10.1183/13993003.00457-2019
Epigenome-wide association study of lung function level and its change
Abstract
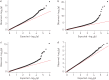
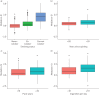
Previous reports link differential DNA methylation (DNAme) to environmental exposures that are associated with lung function. Direct evidence on lung function DNAme is, however, limited. We undertook an agnostic epigenome-wide association study (EWAS) on pre-bronchodilation lung function and its change in adults.In a discovery-replication EWAS design, DNAme in blood and spirometry were measured twice, 6-15 years apart, in the same participants of three adult population-based discovery cohorts (n=2043). Associated DNAme markers (p<5×10-7) were tested in seven replication cohorts (adult: n=3327; childhood: n=420). Technical bias-adjusted residuals of a regression of the normalised absolute β-values on control probe-derived principle components were regressed on level and change of forced expiratory volume in 1 s (FEV1), forced vital capacity (FVC) and their ratio (FEV1/FVC) in the covariate-adjusted discovery EWAS. Inverse-variance-weighted meta-analyses were performed on results from discovery and replication samples in all participants and never-smokers.EWAS signals were enriched for smoking-related DNAme. We replicated 57 lung function DNAme markers in adult, but not childhood samples, all previously associated with smoking. Markers not previously associated with smoking failed replication. cg05575921 (AHRR (aryl hydrocarbon receptor repressor)) showed the statistically most significant association with cross-sectional lung function (FEV1/FVC: pdiscovery=3.96×10-21 and pcombined=7.22×10-50). A score combining 10 DNAme markers previously reported to mediate the effect of smoking on lung function was associated with lung function (FEV1/FVC: p=2.65×10-20).Our results reveal that lung function-associated methylation signals in adults are predominantly smoking related, and possibly of clinical utility in identifying poor lung function and accelerated decline. Larger studies with more repeat time-points are needed to identify lung function DNAme in never-smokers and in children.
Copyright ©ERS 2019.
Conflict of interest statement
Conflict of interest: M. Imboden has nothing to disclose. Conflict of interest: M. Wielscher has nothing to disclose. Conflict of interest: F.I. Rezwan has nothing to disclose. Conflict of interest: A.F.S. Amaral has nothing to disclose. Conflict of interest: E. Schaffner has nothing to disclose. Conflict of interest: A. Jeong has nothing to disclose. Conflict of interest: A. Beckmeyer-Borowko has nothing to disclose. Conflict of interest: S.E. Harris reports grants from Medical Research Council, Biotechnology and Biological Sciences Research Council, Age UK and The Wellcome Trust, during the conduct of the study. Conflict of interest: J.M. Starr has nothing to disclose. Conflict of interest: I.J. Deary reports grants from Age UK and Medical Research Council, during the conduct of the study. Conflict of interest: C. Flexeder has nothing to disclose. Conflict of interest: M. Waldenberger has nothing to disclose. Conflict of interest: A. Peters has nothing to disclose. Conflict of interest: H. Schulz reports grants from German Federal Ministry of Education and Research (BMBF), during the conduct of the study. Conflict of interest: S. Chen has nothing to disclose. Conflict of interest: S.K. Sunny has nothing to disclose. Conflict of interest: W.J.J. Karmaus has nothing to disclose. Conflict of interest: Y. Jiang has nothing to disclose. Conflict of interest: G. Erhart has nothing to disclose. Conflict of interest: F. Kronenberg has nothing to disclose. Conflict of interest: R. Arathimos has nothing to disclose. Conflict of interest: G.C. Sharp has nothing to disclose. Conflict of interest: A.J. Henderson reports grants from Medical Research Council and Wellcome Trust, during the conduct of the study. Conflict of interest: Y. Fu has nothing to disclose. Conflict of interest: P. Piirilä has nothing to disclose. Conflict of interest: K.H. Pietiläinen has nothing to disclose. Conflict of interest: M. Ollikainen has nothing to disclose. Conflict of interest: A. Johansson has nothing to disclose. Conflict of interest: U. Gyllensten has nothing to disclose. Conflict of interest: M de Vries has nothing to disclose. Conflict of interest: D.A. van der Plaat has nothing to disclose. Conflict of interest: K. de Jong has nothing to disclose. Conflict of interest: H.M. Boezen has nothing to disclose. Conflict of interest: I.P. Hall reports grants from GSK and Boehringer Ingelheim, outside the submitted work. Conflict of interest: M.D. Tobin reports grants from Pfizer and GSK, outside the submitted work. Conflict of interest: M-R. Jarvelin has nothing to disclose. Conflict of interest: J.W. Holloway reports grants from European Union and National Institutes of Health, during the conduct of the study. Conflict of interest: D. Jarvis reports grants from European Union, Medical Research Council and Asthma UK, during the conduct of the study. Conflict of interest: N.M. Probst-Hensch has nothing to disclose.
Figures




Comment in
-
Methylation, smoking, and reduced lung function.Eur Respir J. 2019 Jul 4;54(1):1900920. doi: 10.1183/13993003.00920-2019. Print 2019 Jul. Eur Respir J. 2019. PMID: 31273037 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
