A Comprehensive UHPLC Ion Mobility Quadrupole Time-of-Flight Method for Profiling and Quantification of Eicosanoids, Other Oxylipins, and Fatty Acids
- PMID: 31074960
- PMCID: PMC7613057
- DOI: 10.1021/acs.analchem.8b04615
A Comprehensive UHPLC Ion Mobility Quadrupole Time-of-Flight Method for Profiling and Quantification of Eicosanoids, Other Oxylipins, and Fatty Acids
Abstract
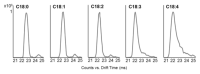
Analysis of oxylipins by liquid chromatography mass spectrometry (LC/MS) is challenging because of the small mass range occupied by this diverse lipid class, the presence of numerous structural isomers, and their low abundance in biological samples. Although highly sensitive LC/MS/MS methods are commonly used, further separation is achievable by using drift tube ion mobility coupled with high-resolution mass spectrometry (DTIM-MS). Herein, we present a combined analytical and computational method for the identification of oxylipins and fatty acids. We use a reversed-phase LC/DTIM-MS workflow able to profile and quantify (based on chromatographic peak area) the oxylipin and fatty acid content of biological samples while simultaneously acquiring full scan and product ion spectra. The information regarding accurate mass, collision-cross-section values in nitrogen (DTCCSN2), and retention times of the species found are compared to an internal library of lipid standards as well as the LIPID MAPS Structure Database by using specifically developed processing tools. Features detected within the DTCCSN2 and m/ z ranges of the analyzed standards are flagged as oxylipin-like species, which can be further characterized using drift-time alignment of product and precursor ions distinctive of DTIM-MS. This not only helps identification by reducing the number of annotations from LIPID MAPS but also guides discovery studies of potentially novel species. Testing the methodology on Salmonella enterica serovar Typhimurium-infected murine bone-marrow-derived macrophages and thrombin activated human platelets yields results in agreement with literature. This workflow has also annotated features as potentially novel oxylipins, confirming its ability in providing further insights into lipid analysis of biological samples.
Figures



References
-
- Smith WL, Murphy RC. The Eicosanoids: Cyclooxygenase, Lipoxygenase, and Epoxygenase Pathways. Biochem Lipids, Lipoproteins Membr. 2002;(4):341–371.
-
- Theken KN, Schuck RN, Edin ML, Tran B, Ellis K, Bass A, Lih FB, Tomer KB, Poloyac SM, Wu MC, et al. Evaluation of Cytochrome P450-Derived Eicosanoids in Humans with Stable Atherosclerotic Cardiovascular Disease. Atherosclerosis. 2012;222(2):530–536. doi: 10.1016/j.atherosclerosis.2012.03.022. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

