Two modes of transvection at the eyes absent gene of Drosophila demonstrate plasticity in transcriptional regulatory interactions in cis and in trans
- PMID: 31075100
- PMCID: PMC6530868
- DOI: 10.1371/journal.pgen.1008152
Two modes of transvection at the eyes absent gene of Drosophila demonstrate plasticity in transcriptional regulatory interactions in cis and in trans
Abstract
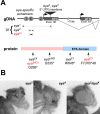
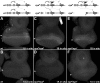
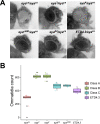
For many genes, proper gene expression requires coordinated and dynamic interactions between multiple regulatory elements, each of which can either promote or silence transcription. In Drosophila, the complexity of the regulatory landscape is further complicated by the tight physical pairing of homologous chromosomes, which can permit regulatory elements to interact in trans, a phenomenon known as transvection. To better understand how gene expression can be programmed through cis- and trans-regulatory interactions, we analyzed transvection effects for a collection of alleles of the eyes absent (eya) gene. We find that trans-activation of a promoter by the eya eye-specific enhancers is broadly supported in many allelic backgrounds, and that the availability of an enhancer to act in trans can be predicted based on the molecular lesion of an eya allele. Furthermore, by manipulating promoter availability in cis and in trans, we demonstrate that the eye-specific enhancers of eya show plasticity in their promoter preference between two different transcriptional start sites, which depends on promoter competition between the two potential targets. Finally, we show that certain alleles of eya demonstrate pairing-sensitive silencing resulting from trans-interactions between Polycomb Response Elements (PREs), and genetic and genomic data support a general role for PcG proteins in mediating transcriptional silencing at eya. Overall, our data highlight how eya gene regulation relies upon a complex but plastic interplay between multiple enhancers, promoters, and PREs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

