RAG2 localization and dynamics in the pre-B cell nucleus
- PMID: 31075127
- PMCID: PMC6510410
- DOI: 10.1371/journal.pone.0216137
RAG2 localization and dynamics in the pre-B cell nucleus
Abstract
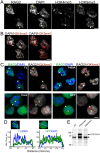
RAG2 of the V(D)J recombinase is essential for lymphocyte development. Within the RAG2 noncore region is a plant homeodomain (PHD) that interacts with the modified histone H3K4me3, and this interaction is important for relieving inhibition of the RAG recombinase for V(D)J recombination. However, the effect of the noncore region on RAG2 localization and dynamics in cell nuclei is poorly understood. Here, we used cell imaging to measure the effect of mutating the RAG2 noncore region on properties of the full length protein. We measured GFP-labeled full length RAG2 (FL), the RAG2 core region alone (Core), and a T490A mutant in the noncore region, which has unique regulatory properties. This showed that FL, T490A, and Core localized to nuclear domains that were adjacent to DAPI-rich heterochromatin, and that contained the active chromatin marker H3K4me3. Within the RAG2-enriched regions, T490A exhibited greater colocalization with H3K4me3 than either FL or Core. Furthermore, colocalization of H3K4me3 with FL and T490A, but not Core, increased in conditions that increased H3K4me3 levels. Superresolution imaging showed H3K4me3 was distributed as puncta that RAG2 abutted, and mobility measurements showed that T490A had a significantly lower rate of diffusion within the nucleus than either FL or Core proteins. Finally, mutating Trp453 of the T490A mutant (W453A,T490A), which blocks PHD-dependent interactions with H3K4me3, abolished the T490A-mediated increased colocalization with H3K4me3 and slower mobility compared to FL. Altogether, these data show that Thr490 in the noncore region modulates RAG2 localization and dynamics in the pre-B cell nucleus, such as by affecting RAG2 interactions with H3K4me3.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

