Identification of Histone H3 (HH3) Genes in Gossypium hirsutum Revealed Diverse Expression During Ovule Development and Stress Responses
- PMID: 31075950
- PMCID: PMC6562411
- DOI: 10.3390/genes10050355
Identification of Histone H3 (HH3) Genes in Gossypium hirsutum Revealed Diverse Expression During Ovule Development and Stress Responses
Abstract
Histone acts as the core for nucleosomes and is a key protein component of chromatin. Among different histone variants, histone H3 (HH3) variants have been reported to play vital roles in plant development. However, biological information and evolutionary relationships of HH3 genes in cotton remain to be elucidated. The current study identified 34 HH3 genes in Gossypium hirsutum. Phylogenetic analysis classified HH3 genes of 19 plant species into eight distinct clades. Sequence logos analysis among Arabidopsis, rice, and G. hirsutum amino acid residues showed higher conservation in amino acids. Using collinearity analysis, we identified 81 orthologous/paralogous gene pairs among the four genomes (A, D, At, and Dt) of cotton. Further, orthologous/paralogous and the Ka/Ks ratio demonstrated that cotton HH3 genes experienced strong purifying selection pressure with restricted functional divergence resulting from segmental and whole genome duplication. Expression pattern analysis indicated that GhHH3 genes were preferentially expressed in cotton ovule tissues. Additionally, GhHH3 gene expression can be regulated by abiotic stresses (cold, heat, sodium chloride (NaCl), and polyethylene glycol (PEG)) and phytohormonal (brassinolide (BL), gibberellic acid (GA), indole-3-acetic acid (IAA), salicylic acid (SA), and methyl jasmonate (MeJA)) treatments, suggesting that GhHH3 genes might play roles in abiotic and hormone stress resistance. Taken together, this work provides important information to decipher complete molecular and physiological functions of HH3 genes in cotton.
Keywords: GhHH3; Gossypium hirsutum; abiotic stress; cis-elements; expression pattern; gene duplication; phylogenetic analysis; phytohormonal stress.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



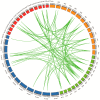

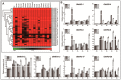
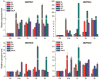

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

