Second-generation molecular subgrouping of medulloblastoma: an international meta-analysis of Group 3 and Group 4 subtypes
- PMID: 31076851
- PMCID: PMC6660496
- DOI: 10.1007/s00401-019-02020-0
Second-generation molecular subgrouping of medulloblastoma: an international meta-analysis of Group 3 and Group 4 subtypes
Abstract
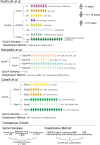
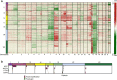
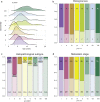
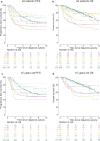
In 2012, an international consensus paper reported that medulloblastoma comprises four molecular subgroups (WNT, SHH, Group 3, and Group 4), each associated with distinct genomic features and clinical behavior. Independently, multiple recent reports have defined further intra-subgroup heterogeneity in the form of biologically and clinically relevant subtypes. However, owing to differences in patient cohorts and analytical methods, estimates of subtype number and definition have been inconsistent, especially within Group 3 and Group 4. Herein, we aimed to reconcile the definition of Group 3/Group 4 MB subtypes through the analysis of a series of 1501 medulloblastomas with DNA-methylation profiling data, including 852 with matched transcriptome data. Using multiple complementary bioinformatic approaches, we compared the concordance of subtype calls between published cohorts and analytical methods, including assessments of class-definition confidence and reproducibility. While the lowest complexity solutions continued to support the original consensus subgroups of Group 3 and Group 4, our analysis most strongly supported a definition comprising eight robust Group 3/Group 4 subtypes (types I-VIII). Subtype II was consistently identified across all component studies, while all others were supported by multiple class-definition methods. Regardless of analytical technique, increasing cohort size did not further increase the number of identified Group 3/Group 4 subtypes. Summarizing the molecular and clinico-pathological features of these eight subtypes indicated enrichment of specific driver gene alterations and cytogenetic events amongst subtypes, and identified highly disparate survival outcomes, further supporting their biological and clinical relevance. Collectively, this study provides continued support for consensus Groups 3 and 4 while enabling robust derivation of, and categorical accounting for, the extensive intertumoral heterogeneity within Groups 3 and 4, revealed by recent high-resolution subclassification approaches. Furthermore, these findings provide a basis for application of emerging methods (e.g., proteomics/single-cell approaches) which may additionally inform medulloblastoma subclassification. Outputs from this study will help shape definition of the next generation of medulloblastoma clinical protocols and facilitate the application of enhanced molecularly guided risk stratification to improve outcomes and quality of life for patients and their families.
Keywords: Biomarkers; Medulloblastoma; Meta-analysis; Methylation; Subtypes.
Figures



References
-
- Archer TC, Ehrenberger T, Mundt F, Gold MP, Krug K, Mah CK, Mahoney EL, Daniel CJ, LeNail A, Ramamoorthy D, et al. Proteomics, post-translational modifications, and integrative analyses reveal molecular heterogeneity within medulloblastoma subgroups. Cancer Cell. 2018;34(3):396–410. doi: 10.1016/j.ccell.2018.08.004. - DOI - PMC - PubMed
-
- Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, Berhoukim R, Amani V, Goumnerova L, Eberhart CG, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. - DOI - PMC - PubMed
-
- Clifford SC, Lusher ME, Lindsey JC, Langdon JA, Gilbertson RJ, Straughton D, Ellison DW. Wnt/Wingless pathway activation and chromosome 6 loss characterize a distinct molecular sub-group of medulloblastomas associated with a favorable prognosis. Cell Cycle. 2006;5:2666–2670. doi: 10.4161/cc.5.22.3446. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

