Exact hypothesis testing for shrinkage-based Gaussian graphical models
- PMID: 31077287
- PMCID: PMC6901079
- DOI: 10.1093/bioinformatics/btz357
Exact hypothesis testing for shrinkage-based Gaussian graphical models
Abstract
Motivation: One of the main goals in systems biology is to learn molecular regulatory networks from quantitative profile data. In particular, Gaussian graphical models (GGMs) are widely used network models in bioinformatics where variables (e.g. transcripts, metabolites or proteins) are represented by nodes, and pairs of nodes are connected with an edge according to their partial correlation. Reconstructing a GGM from data is a challenging task when the sample size is smaller than the number of variables. The main problem consists in finding the inverse of the covariance estimator which is ill-conditioned in this case. Shrinkage-based covariance estimators are a popular approach, producing an invertible 'shrunk' covariance. However, a proper significance test for the 'shrunk' partial correlation (i.e. the GGM edges) is an open challenge as a probability density including the shrinkage is unknown. In this article, we present (i) a geometric reformulation of the shrinkage-based GGM, and (ii) a probability density that naturally includes the shrinkage parameter.
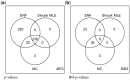
Results: Our results show that the inference using this new 'shrunk' probability density is as accurate as Monte Carlo estimation (an unbiased non-parametric method) for any shrinkage value, while being computationally more efficient. We show on synthetic data how the novel test for significance allows an accurate control of the Type I error and outperforms the network reconstruction obtained by the widely used R package GeneNet. This is further highlighted in two gene expression datasets from stress response in Eschericha coli, and the effect of influenza infection in Mus musculus.
Availability and implementation: https://github.com/V-Bernal/GGM-Shrinkage.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2019. Published by Oxford University Press.
Figures






References
-
- Benjamini Y., Yosef H. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol., 57, 289–300.
-
- Butte A.J., Kohane I.S. (2003) Relevance networks: a first step toward finding genetic regulatory networks within microarray data In: Parmigiani G.et al. (ed.) The Analysis of Gene Expression Data. Springer, New York, NY, pp. 428–446.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

