Gigwa v2-Extended and improved genotype investigator
- PMID: 31077313
- PMCID: PMC6511067
- DOI: 10.1093/gigascience/giz051
Gigwa v2-Extended and improved genotype investigator
Abstract
Background: The study of genetic variations is the basis of many research domains in biology. From genome structure to population dynamics, many applications involve the use of genetic variants. The advent of next-generation sequencing technologies led to such a flood of data that the daily work of scientists is often more focused on data management than data analysis. This mass of genotyping data poses several computational challenges in terms of storage, search, sharing, analysis, and visualization. While existing tools try to solve these challenges, few of them offer a comprehensive and scalable solution.
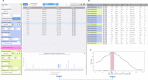
Results: Gigwa v2 is an easy-to-use, species-agnostic web application for managing and exploring high-density genotyping data. It can handle multiple databases and may be installed on a local computer or deployed as an online data portal. It supports various standard import and export formats, provides advanced filtering options, and offers means to visualize density charts or push selected data into various stand-alone or online tools. It implements 2 standard RESTful application programming interfaces, GA4GH, which is health-oriented, and BrAPI, which is breeding-oriented, thus offering wide possibilities of interaction with third-party applications. The project home page provides a list of live instances allowing users to test the system on public data (or reasonably sized user-provided data).
Conclusions: This new version of Gigwa provides a more intuitive and more powerful way to explore large amounts of genotyping data by offering a scalable solution to search for genotype patterns, functional annotations, or more complex filtering. Furthermore, its user-friendliness and interoperability make it widely accessible to the life science community.
Keywords: BrAPI; GA4GH; HapMap; MongoDB; NoSQL; PLINK; REST; SNP; VCF; genomic variations; indel; interoperability; web.
© The Author(s) 2019. Published by Oxford University Press.
Figures


References
-
- Slifer SH. PLINK: key functions for data analysis. Curr Protoc Hum Genet. 2018;97:e59. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

