Evaluation of critical design parameters for RT-qPCR-based analysis of multiple dUTPase isoform genes in mice
- PMID: 31077566
- PMCID: PMC6551494
- DOI: 10.1002/2211-5463.12654
Evaluation of critical design parameters for RT-qPCR-based analysis of multiple dUTPase isoform genes in mice
Abstract
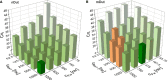
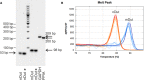
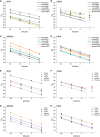
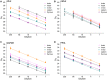
The coupling of nucleotide biosynthesis and genome integrity plays an important role in ensuring faithful maintenance and transmission of genetic information. The enzyme dUTPase is a prime example of such coupling, as it generates dUMP for thymidylate biosynthesis and removes dUTP for synthesis of uracil-free DNA. Despite its significant role, the expression patterns of dUTPase isoforms in animals have not yet been described. Here, we developed a detailed optimization procedure for RT-qPCR-based isoform-specific analysis of dUTPase expression levels in various organs of adult mice. Primer design, optimal annealing temperature, and primer concentrations were specified for both nuclear and mitochondrial dUTPase isoforms, as well as two commonly used reference genes, GAPDH and PPIA. The linear range of the RNA concentration for the reverse transcription reaction was determined. The PCR efficiencies were calculated using serial dilutions of cDNA. Our data indicate that organs involved in lymphocyte production, as well as reproductive organs, are characterized by high levels of expression of the nuclear dUTPase isoform. On the other hand, we observed that expression of the mitochondrial dUTPase isoform is considerably increased in heart, kidney, and ovary. Despite the differences in expression levels among the various organs, we also found that the mitochondrial dUTPase isoform shows a much more uniform expression pattern as compared to the reference genes GAPDH and PPIA.
Keywords: RT-qPCR optimization; dUTPase; isoform-specific expression levels; mitochondrial dUTPase isoform; mouse organs; nuclear dUTPase isoform.
© 2019 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Kumar H, Kehrer J, Singer M, Reinig M, Santos JM, Mair GR and Frischknecht F (2019) Functional genetic evaluation of DNA house‐cleaning enzymes in the malaria parasite: dUTPase and Ap4AH are essential in Plasmodium berghei but ITPase and NDH are dispensable. Expert Opin Ther Targets 23, 251–261. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

