Structural basis for catalysis and substrate specificity of a 3C-like cysteine protease from a mosquito mesonivirus
- PMID: 31078932
- PMCID: PMC7111312
- DOI: 10.1016/j.virol.2019.05.001
Structural basis for catalysis and substrate specificity of a 3C-like cysteine protease from a mosquito mesonivirus
Abstract
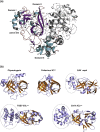
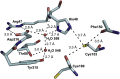
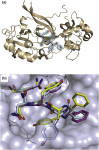
Cavally virus (CavV) is a mosquito-borne plus-strand RNA virus in the family Mesoniviridae (order Nidovirales). We present X-ray structures for the CavV 3C-like protease (3CLpro), as a free enzyme and in complex with a peptide aldehyde inhibitor mimicking the P4-to-P1 residues of a natural substrate. The 3CLpro structure (refined to 1.94 Å) shows that the protein forms dimers. The monomers are comprised of N-terminal domains I and II, which adopt a chymotrypsin-like fold, and a C-terminal α-helical domain III. The catalytic Cys-His dyad is assisted by a complex network of interactions involving a water molecule that mediates polar contacts between the catalytic His and a conserved Asp located in the domain II-III junction and is suitably positioned to stabilize the developing positive charge of the catalytic His in the transition state during catalysis. The study also reveals the structural basis for the distinct P2 Asn-specific substrate-binding pocket of mesonivirus 3CLpros.
Keywords: 3C-like protease; Active site of chymotrypsin-like proteases; Coronavirus; Crystal structure; Invertebrate RNA virus; Mesonivirus.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., Richardson J.S., Terwilliger T.C., Zwart P.H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. - PMC - PubMed
-
- Adams M.J., Lefkowitz E.J., King A.M., Harrach B., Harrison R.L., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Mushegian A.R., Nibert M., Sabanadzovic S., Sanfacon H., Siddell S.G., Simmonds P., Varsani A., Zerbini F.M., Gorbalenya A.E., Davison A.J. Ratification vote on taxonomic proposals to the international committee on taxonomy of viruses (2016) Arch. Virol. 2016;161:2921–2949. - PMC - PubMed
-
- Allaire M., Chernaia M.M., Malcolm B.A., James M.N. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature. 1994;369:72–76. - PubMed
-
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

