Slow Delivery Immunization Enhances HIV Neutralizing Antibody and Germinal Center Responses via Modulation of Immunodominance
- PMID: 31080066
- PMCID: PMC6619430
- DOI: 10.1016/j.cell.2019.04.012
Slow Delivery Immunization Enhances HIV Neutralizing Antibody and Germinal Center Responses via Modulation of Immunodominance
Erratum in
-
Slow Delivery Immunization Enhances HIV Neutralizing Antibody and Germinal Center Responses via Modulation of Immunodominance.Cell. 2020 Jan 9;180(1):206. doi: 10.1016/j.cell.2019.12.027. Cell. 2020. PMID: 31923396 Free PMC article. No abstract available.
Abstract
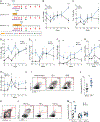
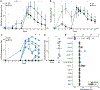
Conventional immunization strategies will likely be insufficient for the development of a broadly neutralizing antibody (bnAb) vaccine for HIV or other difficult pathogens because of the immunological hurdles posed, including B cell immunodominance and germinal center (GC) quantity and quality. We found that two independent methods of slow delivery immunization of rhesus monkeys (RMs) resulted in more robust T follicular helper (TFH) cell responses and GC B cells with improved Env-binding, tracked by longitudinal fine needle aspirates. Improved GCs correlated with the development of >20-fold higher titers of autologous nAbs. Using a new RM genomic immunoglobulin locus reference, we identified differential IgV gene use between immunization modalities. Ab mapping demonstrated targeting of immunodominant non-neutralizing epitopes by conventional bolus-immunized animals, whereas slow delivery-immunized animals targeted a more diverse set of epitopes. Thus, alternative immunization strategies can enhance nAb development by altering GCs and modulating the immunodominance of non-neutralizing epitopes.
Keywords: FNA; GC- T(FH); HIV vaccine; affinity maturation; immune complexes; memory B cells; non-human primates; rhesus macaque genome; somatic hypermutation.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures





References
-
- Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, Kulp DW, Bhullar D, Kalyuzhniy O, Havenar-Daughton C, et al. (2018). Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity 48, 133–146.e136. - PMC - PubMed
-
- Andrew S (2010). FastQC: A quality control tool for high throughput sequence data. 2010.
-
- Andrews SF, Graham BS, Mascola JR, and McDermott AB (2018). Is It Possible to Develop a “Universal” Influenza Virus Vaccine? Immunogenetic Considerations Underlying B-Cell Biology in he Development of a Pan-Subtype Influenza A Vaccine Targeting the Hemagglutinin Stem. Cold Spring Harb Perspect Biol 10, a029413. - PMC - PubMed
-
- Angeletti D, and Yewdell JW (2018). Understanding and Manipulating Viral Immunity: Antibody Immunodominance Enters Center Stage. Trends Immunol 39, 549–561. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI124796/AI/NIAID NIH HHS/United States
- P51 RR000165/RR/NCRR NIH HHS/United States
- R01 AI136621/AI/NIAID NIH HHS/United States
- F31 AI131873/AI/NIAID NIH HHS/United States
- UM1 AI124436/AI/NIAID NIH HHS/United States
- U24 AI120134/AI/NIAID NIH HHS/United States
- UM1 AI100663/AI/NIAID NIH HHS/United States
- P30 NS072030/NS/NINDS NIH HHS/United States
- P51 OD011132/OD/NIH HHS/United States
- R01 AI125068/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R37 AI125068/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

