Progression of the canonical reference malaria parasite genome from 2002-2019
- PMID: 31080894
- PMCID: PMC6484455
- DOI: 10.12688/wellcomeopenres.15194.2
Progression of the canonical reference malaria parasite genome from 2002-2019
Abstract
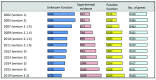
Here we describe the ways in which the sequence and annotation of the Plasmodium falciparum reference genome has changed since its publication in 2002. As the malaria species responsible for the most deaths worldwide, the richness of annotation and accuracy of the sequence are important resources for the P. falciparum research community as well as the basis for interpreting the genomes of subsequently sequenced species. At the time of publication in 2002 over 60% of predicted genes had unknown functions. As of March 2019, this number has been significantly decreased to 33%. The reduction is due to the inclusion of genes that were subsequently characterised experimentally and genes with significant similarity to others with known functions. In addition, the structural annotation of genes has been significantly refined; 27% of gene structures have been changed since 2002, comprising changes in exon-intron boundaries, addition or deletion of exons and the addition or deletion of genes. The sequence has also undergone significant improvements. In addition to the correction of a large number of single-base and insertion or deletion errors, a major miss-assembly between the subtelomeres of chromosome 7 and 8 has been corrected. As the number of sequenced isolates continues to grow rapidly, a single reference genome will not be an adequate basis for interpreting intra-species sequence diversity. We therefore describe in this publication a population reference genome of P. falciparum, called Pfref1. This reference will enable the community to map to regions that are not present in the current assembly. P. falciparum 3D7 will continue to be maintained, with ongoing curation ensuring continual improvements in annotation quality.
Keywords: Plasmodium; annotation; curation; falciparum; genome; reference.
Conflict of interest statement
No competing interests were disclosed.
Figures





References
-
- Böhme U: Progression of the canonical reference malaria parasite genome from 2002–2019.2019. 10.17605/OSF.IO/5K9VJ - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

