Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine
- PMID: 31081034
- PMCID: PMC6895265
- DOI: 10.1093/nar/gkz381
Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine
Abstract
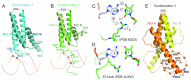
The psychiatric risk-associated transcription factor 4 (TCF4) is linked to schizophrenia. Rare TCF4 coding variants are found in individuals with Pitt-Hopkins syndrome-an intellectual disability and autism spectrum disorder. TCF4 contains a C-terminal basic-helix-loop-helix (bHLH) DNA binding domain which recognizes the enhancer-box (E-box) element 5'-CANNTG-3' (where N = any nucleotide). A subset of the TCF4-occupancy sites have the expanded consensus binding specificity 5'-C(A/G)-CANNTG-3', with an added outer Cp(A/G) dinucleotide; for example in the promoter for CNIH3, a gene involved in opioid dependence. In mammalian genomes, particularly brain, the CpG and CpA dinucleotides can be methylated at the 5-position of cytosine (5mC), and then may undergo successive oxidations to the 5-hydroxymethyl (5hmC), 5-formyl (5fC), and 5-carboxyl (5caC) forms. We find that, in the context of 5'-0CG-1CA-2CG-3TG-3'(where the numbers indicate successive dinucleotides), modification of the central E-box 2CG has very little effect on TCF4 binding, E-box 1CA modification has a negative influence on binding, while modification of the flanking 0CG, particularly carboxylation, has a strong positive impact on TCF4 binding to DNA. Crystallization of TCF4 in complex with unmodified or 5caC-modified oligonucleotides revealed that the basic region of bHLH domain adopts multiple conformations, including an extended loop going through the DNA minor groove, or the N-terminal portion of a long helix binding in the DNA major groove. The different protein conformations enable arginine 576 (R576) to interact, respectively, with a thymine in the minor groove, a phosphate group of DNA backbone, or 5caC in the major groove. The Pitt-Hopkins syndrome mutations affect five arginine residues in the basic region, two of them (R569 and R576) involved in 5caC recognition. Our analyses indicate, and suggest a structural basis for, the preferential recognition of 5caC by a transcription factor centrally important in brain development.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Forrest M.P., Hill M.J., Quantock A.J., Martin-Rendon E., Blake D.J.. The emerging roles of TCF4 in disease and development. Trends Mol. Med. 2014; 20:322–331. - PubMed
-
- Caudy M., Vassin H., Brand M., Tuma R., Jan L.Y., Jan Y.N.. daughterless, a Drosophila gene essential for both neurogenesis and sex determination, has sequence similarities to myc and the achaete-scute complex. Cell. 1988; 55:1061–1067. - PubMed
-
- Murre C., McCaw P.S., Baltimore D.. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989; 56:777–783. - PubMed
-
- Henthorn P., Kiledjian M., Kadesch T.. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science. 1990; 247:467–470. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

