AnnoTree: visualization and exploration of a functionally annotated microbial tree of life
- PMID: 31081040
- PMCID: PMC6511854
- DOI: 10.1093/nar/gkz246
AnnoTree: visualization and exploration of a functionally annotated microbial tree of life
Abstract
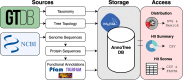
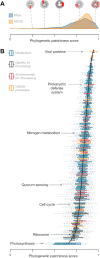
Bacterial genomics has revolutionized our understanding of the microbial tree of life; however, mapping and visualizing the distribution of functional traits across bacteria remains a challenge. Here, we introduce AnnoTree-an interactive, functionally annotated bacterial tree of life that integrates taxonomic, phylogenetic and functional annotation data from over 27 000 bacterial and 1500 archaeal genomes. AnnoTree enables visualization of millions of precomputed genome annotations across the bacterial and archaeal phylogenies, thereby allowing users to explore gene distributions as well as patterns of gene gain and loss in prokaryotes. Using AnnoTree, we examined the phylogenomic distributions of 28 311 gene/protein families, and measured their phylogenetic conservation, patchiness, and lineage-specificity within bacteria. Our analyses revealed widespread phylogenetic patchiness among bacterial gene families, reflecting the dynamic evolution of prokaryotic genomes. Genes involved in phage infection/defense, mobile elements, and antibiotic resistance dominated the list of most patchy traits, as well as numerous intriguing metabolic enzymes that appear to have undergone frequent horizontal transfer. We anticipate that AnnoTree will be a valuable resource for exploring prokaryotic gene histories, and will act as a catalyst for biological and evolutionary hypothesis generation. AnnoTree is freely available at http://annotree.uwaterloo.ca.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Venter J.C., Remington K., Heidelberg J.F., Halpern A.L., Rusch D., Eisen Ja., Wu D., Paulsen I., Nelson K.E., Nelson W. et al.. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004; 304:66–74. - PubMed
-
- Demuth J., Hahn M.. The life and death of gene families. Bioessays. 2009; 31:29–39. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

