History of the Selective Autophagy Research: How Did It Begin and Where Does It Stand Today?
- PMID: 31082435
- PMCID: PMC6971693
- DOI: 10.1016/j.jmb.2019.05.010
History of the Selective Autophagy Research: How Did It Begin and Where Does It Stand Today?
Abstract
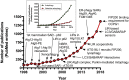
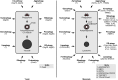
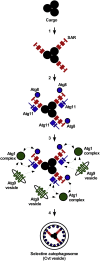
Autophagy, self-eating, is a pivotal catabolic mechanism that ensures homeostasis and survival of the cell in the face of stressors as different as starvation, infection, or protein misfolding. The importance of the research in this field was recognized by the general public after the Nobel Prize for Physiology or Medicine was awarded in 2016 to Yoshinori Ohsumi for discoveries of the mechanisms of autophagy using yeast as a model organism. One of the seminal findings of Ohsumi was on the role ubiquitin-like proteins (UBLs)-Atg5, Atg12, and Atg8-play in the formation of the double-membrane vesicle autophagosome, which is the functional unit of autophagy. Subsequent work by several groups demonstrated that, like the founding member of the UBL family ubiquitin, these small but versatile protein and lipid modifiers interact with a plethora of proteins, which either directly regulate autophagosome formation, for example, components of the Atg1/ULK1 complex, or are involved in cargo recognition, for example, Atg19 and p62/SQSTM1. By tethering the cargo to the UBLs present on the forming autophagosome, the latter proteins were proposed to effectively act as selective autophagy receptors. The discovery of the selective autophagy receptors brought a breakthrough in the autophagy field, supplying the mechanistic underpinning for the formation of an autophagosome selectively around the cytosolic cargo, that is, a protein aggregate, a mitochondrion, or a cytosolic bacterium. In this historical overview, I highlight key steps that the research into selective autophagy has been taking over the past 20 years. I comment on their significance and discuss current challenges in developing more detailed knowledge of the mechanisms of selective autophagy. I will conclude by introducing the new directions that this dynamic research field is taking into its third decade.
Keywords: Atg8; GABARAP; LC3; SAR; UBL.
Copyright © 2019 The Author. Published by Elsevier Ltd.. All rights reserved.
Figures


References
-
- Klionsky D.J. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. - PubMed
-
- Tsukada M., Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. - PubMed
-
- Thumm M., Egner R., Koch B., Schlumpberger M., Straub M., Veenhuis M. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

