The Role of Archaeal Chromatin in Transcription
- PMID: 31082442
- PMCID: PMC6842674
- DOI: 10.1016/j.jmb.2019.05.006
The Role of Archaeal Chromatin in Transcription
Abstract
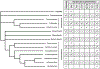
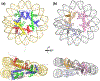
Genomic organization impacts accessibility and movement of information processing systems along DNA. DNA-bound proteins dynamically dictate gene expression and provide regulatory potential to tune transcription rates to match ever-changing environmental conditions. Archaeal genomes are typically small, circular, gene dense, and organized either by histone proteins that are homologous to their eukaryotic counterparts, or small basic proteins that function analogously to bacterial nucleoid proteins. We review here how archaeal genomes are organized and how such organization impacts archaeal gene expression, focusing on conserved DNA-binding proteins within the clade and the factors that are known to impact transcription initiation and elongation within protein-bound genomes.
Keywords: Alba; RNA polymerase; archaea; histone; transcription regulation.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Figures
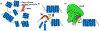
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

