A Symphony of Signals: Intercellular and Intracellular Signaling Mechanisms Underlying Circadian Timekeeping in Mice and Flies
- PMID: 31086044
- PMCID: PMC6540063
- DOI: 10.3390/ijms20092363
A Symphony of Signals: Intercellular and Intracellular Signaling Mechanisms Underlying Circadian Timekeeping in Mice and Flies
Abstract
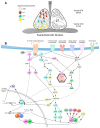
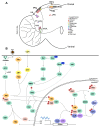
The central pacemakers of circadian timekeeping systems are highly robust yet adaptable, providing the temporal coordination of rhythms in behavior and physiological processes in accordance with the demands imposed by environmental cycles. These features of the central pacemaker are achieved by a multi-oscillator network in which individual cellular oscillators are tightly coupled to the environmental day-night cycle, and to one another via intercellular coupling. In this review, we will summarize the roles of various neurotransmitters and neuropeptides in the regulation of circadian entrainment and synchrony within the mammalian and Drosophila central pacemakers. We will also describe the diverse functions of protein kinases in the relay of input signals to the core oscillator or the direct regulation of the molecular clock machinery.
Keywords: Drosophila; central pacemaker; circadian rhythms; entrainment; intercellular and intracellular signaling; neuropeptides; neurotransmitters; protein kinases; suprachiasmatic nucleus; synchrony.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

