The structural basis of translational control by eIF2 phosphorylation
- PMID: 31086188
- PMCID: PMC6513899
- DOI: 10.1038/s41467-019-10167-3
The structural basis of translational control by eIF2 phosphorylation
Abstract
Protein synthesis in eukaryotes is controlled by signals and stresses via a common pathway, called the integrated stress response (ISR). Phosphorylation of the translation initiation factor eIF2 alpha at a conserved serine residue mediates translational control at the ISR core. To provide insight into the mechanism of translational control we have determined the structures of eIF2 both in phosphorylated and unphosphorylated forms bound with its nucleotide exchange factor eIF2B by electron cryomicroscopy. The structures reveal that eIF2 undergoes large rearrangements to promote binding of eIF2α to the regulatory core of eIF2B comprised of the eIF2B alpha, beta and delta subunits. Only minor differences are observed between eIF2 and eIF2αP binding to eIF2B, suggesting that the higher affinity of eIF2αP for eIF2B drives translational control. We present a model for controlled nucleotide exchange and initiator tRNA binding to the eIF2/eIF2B complex.
Conflict of interest statement
The authors declare no competing interests.
Figures



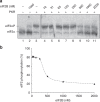
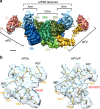

References
-
- Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Wellcome Trust/United Kingdom
- BB/L020157/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/N014049/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)/International
- BB/M006565/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

