EGFR mutation subtypes and response to immune checkpoint blockade treatment in non-small-cell lung cancer
- PMID: 31086949
- PMCID: PMC6683857
- DOI: 10.1093/annonc/mdz141
EGFR mutation subtypes and response to immune checkpoint blockade treatment in non-small-cell lung cancer
Abstract
Background: Although EGFR mutant tumors exhibit low response rates to immune checkpoint blockade overall, some EGFR mutant tumors do respond to these therapies; however, there is a lack of understanding of the characteristics of EGFR mutant lung tumors responsive to immune checkpoint blockade.
Patients and methods: We retrospectively analyzed de-identified clinical and molecular data on 171 cases of EGFR mutant lung tumors treated with immune checkpoint inhibitors from the Yale Cancer Center, Memorial Sloan Kettering Cancer Center, University of California Los Angeles, and Dana Farber Cancer Institute. A separate cohort of 383 EGFR mutant lung cancer cases with sequencing data available from the Yale Cancer Center, Memorial Sloan Kettering Cancer Center, and The Cancer Genome Atlas was compiled to assess the relationship between tumor mutation burden and specific EGFR alterations.
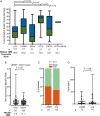
Results: Compared with 212 EGFR wild-type lung cancers, outcomes with programmed cell death 1 or programmed death-ligand 1 (PD-(L)1) blockade were worse in patients with lung tumors harboring alterations in exon 19 of EGFR (EGFRΔ19) but similar for EGFRL858R lung tumors. EGFRT790M status and PD-L1 expression did not impact response or survival outcomes to immune checkpoint blockade. PD-L1 expression was similar across EGFR alleles. Lung tumors with EGFRΔ19 alterations harbored a lower tumor mutation burden compared with EGFRL858R lung tumors despite similar smoking history.
Conclusions: EGFR mutant tumors have generally low response to immune checkpoint inhibitors, but outcomes vary by allele. Understanding the heterogeneity of EGFR mutant tumors may be informative for establishing the benefits and uses of PD-(L)1 therapies for patients with this disease.
Keywords: epidermal growth factor receptor; immune checkpoint blockade; non-small-cell lung cancer.
© The Author(s) 2019. Published by Oxford University Press on behalf of the European Society for Medical Oncology.
Figures


Comment in
-
EGFR mutation subtype impacts efficacy of immune checkpoint inhibitors in non-small-cell lung cancer.Ann Oncol. 2019 Aug 1;30(8):1190-1192. doi: 10.1093/annonc/mdz185. Ann Oncol. 2019. PMID: 31198952 No abstract available.
-
EGFR mutations in lung cancer: not all equal in the eyes of the immune system?Ann Transl Med. 2019 Dec;7(Suppl 8):S326. doi: 10.21037/atm.2019.09.132. Ann Transl Med. 2019. PMID: 32016044 Free PMC article. No abstract available.
References
-
- Lynch TJ, Bell DW, Sordella R. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004; 350(21): 2129–2139. - PubMed
-
- Paez JG, Janne PA, Lee JC. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004; 304(5676): 1497–1500. - PubMed
-
- Maemondo M, Inoue A, Kobayashi K. et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010; 362(25): 2380–2388. - PubMed
-
- Mitsudomi T, Morita S, Yatabe Y. et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010; 11(2): 121–128. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

